Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil
- PMID: 33605869
- PMCID: PMC8084516
- DOI: 10.3201/eid2705.210191
Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil
Abstract
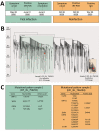
Uncertainty remains about how long the protective immune responses against severe acute respiratory syndrome coronavirus 2 persists, and suspected reinfection in recovered patients has been reported. We describe a case of reinfection from distinct virus lineages in Brazil harboring the E484K mutation, a variant associated with escape from neutralizing antibodies.
Keywords: Brazil; COVID-19; E484K spike mutation; SARS-CoV-2; coronavirus disease; genomic surveillance; reinfection; respiratory infections; severe acute respiratory syndrome coronavirus 2; viruses; zoonoses.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

