Antidepressant drugs act by directly binding to TRKB neurotrophin receptors
- PMID: 33606976
- PMCID: PMC7938888
- DOI: 10.1016/j.cell.2021.01.034
Antidepressant drugs act by directly binding to TRKB neurotrophin receptors
Abstract
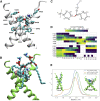
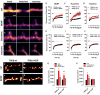
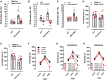
It is unclear how binding of antidepressant drugs to their targets gives rise to the clinical antidepressant effect. We discovered that the transmembrane domain of tyrosine kinase receptor 2 (TRKB), the brain-derived neurotrophic factor (BDNF) receptor that promotes neuronal plasticity and antidepressant responses, has a cholesterol-sensing function that mediates synaptic effects of cholesterol. We then found that both typical and fast-acting antidepressants directly bind to TRKB, thereby facilitating synaptic localization of TRKB and its activation by BDNF. Extensive computational approaches including atomistic molecular dynamics simulations revealed a binding site at the transmembrane region of TRKB dimers. Mutation of the TRKB antidepressant-binding motif impaired cellular, behavioral, and plasticity-promoting responses to antidepressants in vitro and in vivo. We suggest that binding to TRKB and allosteric facilitation of BDNF signaling is the common mechanism for antidepressant action, which may explain why typical antidepressants act slowly and how molecular effects of antidepressants are translated into clinical mood recovery.
Keywords: BDNF; antidepressant; cholesterol; fluoxetine; ketamine; molecular dynamic simulation; neurotrophin; plasticity.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.C. and M.S. are shareholders of Herantis Pharma PIc that is not related to this study. E.C. has received lecture fees from Janssen-Cilag. Other authors declare no conflicts of interest.
Figures


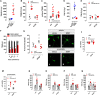
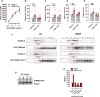

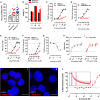

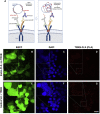


Comment in
-
Fast-Trk(B)ing the mechanism of antidepressants.Neuron. 2021 May 19;109(10):1593-1595. doi: 10.1016/j.neuron.2021.04.027. Epub 2021 May 12. Neuron. 2021. PMID: 34015266
-
Membrane molecules for mood.Trends Neurosci. 2021 Aug;44(8):602-604. doi: 10.1016/j.tins.2021.05.001. Epub 2021 May 27. Trends Neurosci. 2021. PMID: 34053679
References
-
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25.
-
- Ampuero E., Stehberg J., Gonzalez D., Besser N., Ferrero M., Diaz-Veliz G., Wyneken U., Rubio F.J. Repetitive fluoxetine treatment affects long-term memories but not learning. Behav. Brain Res. 2013;247:92–100. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

