Rbbp4 Suppresses Premature Differentiation of Embryonic Stem Cells
- PMID: 33606987
- PMCID: PMC7940252
- DOI: 10.1016/j.stemcr.2021.01.009
Rbbp4 Suppresses Premature Differentiation of Embryonic Stem Cells
Abstract
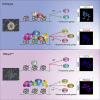
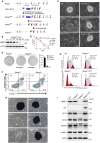
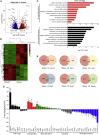
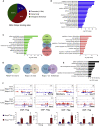
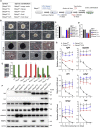
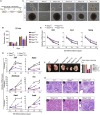
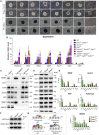
Polycomb group (PcG) proteins exist in distinct multi-protein complexes and play a central role in silencing developmental genes, yet the underlying mechanisms remain elusive. Here, we show that deficiency of retinoblastoma binding protein 4 (RBBP4), a component of the Polycomb repressive complex 2 (PRC2), in embryonic stem cells (ESCs) leads to spontaneous differentiation into mesendodermal lineages. We further show that Rbbp4 and core PRC2 share an important number of common genomic targets, encoding regulators involved in early germ layer specification. Moreover, we find that Rbbp4 is absolutely essential for genomic targeting of PRC2 to a subset of developmental genes. Interestingly, we demonstrate that Rbbp4 is necessary for sustaining the expression of Oct4 and Sox2 and that the forced co-expression of Oct4 and Sox2 fully rescues the pluripotency of Rbbp4-null ESCs. Therefore, our study indicates that Rbbp4 links maintenance of the pluripotency regulatory network with repression of mesendoderm lineages.
Keywords: Oct4; PRC2; RBBP4; Sox2; embryonic stem cells; mesendoderm; pluripotency; polycomb; self-renewal.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Conklin J.F., Baker J., Sage J. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 2012;3:1244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

