Distinct actin-dependent nanoscale assemblies underlie the dynamic and hierarchical organization of E-cadherin
- PMID: 33607036
- PMCID: PMC7611338
- DOI: 10.1016/j.cub.2021.01.059
Distinct actin-dependent nanoscale assemblies underlie the dynamic and hierarchical organization of E-cadherin
Abstract
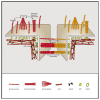
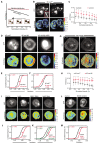
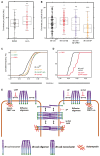
Cadherins are transmembrane adhesion proteins required for the formation of cohesive tissues.1-4 Intracellular interactions of E-cadherin with the Catenin family proteins, α- and β-catenin, facilitate connections with the cortical actomyosin network. This is necessary for maintaining the integrity of cell-cell adhesion in epithelial tissues.5-11 The supra-molecular architecture of E-cadherin is an important feature of its adhesion function; cis and trans interactions of E-cadherin are deployed12-15 to form clusters, both in cis and trans.11,16-21 Studies in Drosophila embryo have also shown that Drosophila E-cadherin (dE-cad) is organized as finite-sized dynamic clusters that localize with actin patches at cell-cell junctions, in continuous exchange with the extra-junctional pool of dE-cad surrounding the clusters.11,19 Here, we use the ectopic expression of dE-cad in larval hemocytes, which lack endogenous dE-cad to recapitulate functional cell-cell junctions in a convenient model system. We find that, while dE-cad at cell-cell junctions in hemocytes exhibits a clustered trans-paired organization similar to that reported previously in embryonic epithelial tissue, extra-junctional dE-cad is also organized as relatively immobile nanoclusters as well as more loosely packed diffusive oligomers. Oligomers are promoted by cis interactions of the ectodomain, and their growth is counteracted by the activity of cortical actomyosin. Oligomers in turn promote assembly of dense nanoclusters that require cortical actomyosin activity. Thus, cortical actin activity remodels oligomers and generates nanoclusters. The requirement for dynamic actin in the organization of dE-cad at the nanoscale may provide a mechanism to dynamically tune junctional strength.
Keywords: E-cadherin; Emmission anisotropy; FCS; actomyosin; adhesion independent nanoscale assembly; cell adhesion; homo-FRET; nanoclusters; oligomers.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Oda H, Tsukita S, Takeichi M. Dynamic behavior of the cad-herin-based cell-cell adhesion system during Drosophila gastrulation. Dev Biol. 1998;203:435–450. - PubMed
-
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Török T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. - PubMed
-
- Uemura T, Oda H, Kraut R, Hayashi S, Kotaoka Y, Takeichi M. Zygotic Drosophila E-cadherin expression is required for processes of dynamic epithelial cell rearrangement in the Drosophila embryo. Genes Dev. 1996;10:659–671. - PubMed
-
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

