Loss of nuclear UBE3A activity is the predominant cause of Angelman syndrome in individuals carrying UBE3A missense mutations
- PMID: 33607653
- PMCID: PMC8101352
- DOI: 10.1093/hmg/ddab050
Loss of nuclear UBE3A activity is the predominant cause of Angelman syndrome in individuals carrying UBE3A missense mutations
Abstract
Angelman syndrome (AS) is a severe neurodevelopmental disorder caused by deletion (~75%) or mutation (~10%) of the ubiquitin E3 ligase A (UBE3A) gene, which encodes a HECT type E3 ubiquitin protein ligase. Although the critical substrates of UBE3A are unknown, previous studies have suggested a critical role of nuclear UBE3A in AS pathophysiology. Here, we investigated to what extent UBE3A missense mutations disrupt UBE3A subcellular localization as well as catalytic activity, stability and protein folding. Our functional screen of 31 UBE3A missense mutants revealed that UBE3A mislocalization is the predominant cause of UBE3A dysfunction, accounting for 55% of the UBE3A mutations tested. The second major cause (29%) is a loss of E3-ubiquitin ligase activity, as assessed in an Escherichia coli in vivo ubiquitination assay. Mutations affecting catalytic activity are found not only in the catalytic HECT domain, but also in the N-terminal half of UBE3A, suggesting an important contribution of this N-terminal region to its catalytic potential. Together, our results show that loss of nuclear UBE3A E3 ligase activity is the predominant cause of UBE3A-linked AS. Moreover, our functional analysis screen allows rapid assessment of the pathogenicity of novel UBE3A missense variants which will be of particular importance when treatments for AS become available.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

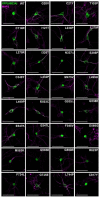
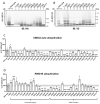


References
-
- Buiting, K., Williams, C. and Horsthemke, B. (2016) Angelman syndrome — insights into a rare neurogenetic disorder. Nat. Rev. Neurol., 12, 584–593. - PubMed
-
- Matsuura, T., Sutcliffe, J.S., Fang, P., Galjaard, R.J., Jiang, Y.-H., Benton, C.S., Rommens, J.M. and Beaudet, A.L. (1997) De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet., 15, 74–77. - PubMed
-
- Kishino, T., Lalande, M. and Wagstaff, J. (1997) UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet., 15, 70–73. - PubMed
-
- Scheffner, M. and Kumar, S. (2014) Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim. Biophys. Acta, 1843, 61–74. - PubMed
-
- Dindot, S.V., Antalffy, B.A., Bhattacharjee, M.B. and Beaudet, A.L. (2008) The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum. Mol. Genet., 17, 111–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

