Performance of model-based multifactor dimensionality reduction methods for epistasis detection by controlling population structure
- PMID: 33608043
- PMCID: PMC7893746
- DOI: 10.1186/s13040-021-00247-w
Performance of model-based multifactor dimensionality reduction methods for epistasis detection by controlling population structure
Abstract
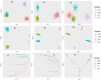
Background: In genome-wide association studies the extent and impact of confounding due to population structure have been well recognized. Inadequate handling of such confounding is likely to lead to spurious associations, hampering replication, and the identification of causal variants. Several strategies have been developed for protecting associations against confounding, the most popular one is based on Principal Component Analysis. In contrast, the extent and impact of confounding due to population structure in gene-gene interaction association epistasis studies are much less investigated and understood. In particular, the role of nonlinear genetic population substructure in epistasis detection is largely under-investigated, especially outside a regression framework.
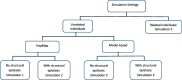
Methods: To identify causal variants in synergy, to improve interpretability and replicability of epistasis results, we introduce three strategies based on a model-based multifactor dimensionality reduction approach for structured populations, namely MBMDR-PC, MBMDR-PG, and MBMDR-GC.
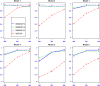
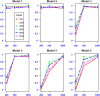
Results: Simulation results comparing the performance of various approaches show that in the presence of population structure MBMDR-PC and MBMDR-PG consistently better control type I error rate at the nominal level than MBMDR-GC. Moreover, our proposed three methods of population structure correction outperform MDR-SP in terms of statistical power.
Conclusion: We demonstrate through extensive simulation studies the effect of various degrees of genetic population structure and relatedness on epistasis detection and propose appropriate remedial measures based on linear and nonlinear sample genetic similarity.
Keywords: Confounding; Epistasis; GWAIS; GWAS; Gene-gene interaction; MB-MDR; Population stratification; Population structure; Principal components.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Simpson EH. The interpretation of interaction in contingency tables. J R Stat Soc Ser B Methodol. 1951;13:238–241.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

