Exploring beyond clinical routine SARS-CoV-2 serology using MultiCoV-Ab to evaluate endemic coronavirus cross-reactivity
- PMID: 33608538
- PMCID: PMC7896075
- DOI: 10.1038/s41467-021-20973-3
Exploring beyond clinical routine SARS-CoV-2 serology using MultiCoV-Ab to evaluate endemic coronavirus cross-reactivity
Abstract
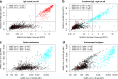
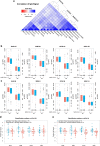
The humoral immune response to SARS-CoV-2 is a benchmark for immunity and detailed analysis is required to understand the manifestation and progression of COVID-19, monitor seroconversion within the general population, and support vaccine development. The majority of currently available commercial serological assays only quantify the SARS-CoV-2 antibody response against individual antigens, limiting our understanding of the immune response. To overcome this, we have developed a multiplex immunoassay (MultiCoV-Ab) including spike and nucleocapsid proteins of SARS-CoV-2 and the endemic human coronaviruses. Compared to three broadly used commercial in vitro diagnostic tests, our MultiCoV-Ab achieves a higher sensitivity and specificity when analyzing a well-characterized sample set of SARS-CoV-2 infected and uninfected individuals. We find a high response against endemic coronaviruses in our sample set, but no consistent cross-reactive IgG response patterns against SARS-CoV-2. Here we show a robust, high-content-enabled, antigen-saving multiplex assay suited to both monitoring vaccination studies and facilitating epidemiologic screenings for humoral immunity towards pandemic and endemic coronaviruses.
Conflict of interest statement
The authors declare the following competing interests: T.O.J. is a scientific advisor for Luminex. N.S-M. was a speaker at Luminex user meetings in the past. The Natural and Medical Sciences Institute at the University of Tübingen is involved in applied research projects as a fee for services with Luminex. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

