Defining the epitope of a blood-brain barrier crossing single domain antibody specific for the type 1 insulin-like growth factor receptor
- PMID: 33608571
- PMCID: PMC7896052
- DOI: 10.1038/s41598-021-83198-w
Defining the epitope of a blood-brain barrier crossing single domain antibody specific for the type 1 insulin-like growth factor receptor
Abstract
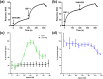
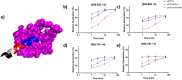
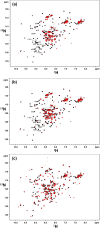
Ligand-activated signaling through the type 1 insulin-like growth factor receptor (IGF1R) is implicated in many physiological processes ranging from normal human growth to cancer proliferation and metastasis. IGF1R has also emerged as a target for receptor-mediated transcytosis, a transport phenomenon that can be exploited to shuttle biotherapeutics across the blood-brain barrier (BBB). We employed differential hydrogen-deuterium exchange mass spectrometry (HDX-MS) and nuclear magnetic resonance (NMR) to characterize the interactions of the IGF1R ectodomain with a recently discovered BBB-crossing single-domain antibody (sdAb), VHH-IR5, in comparison with IGF-1 binding. HDX-MS confirmed that IGF-1 induced global conformational shifts in the L1/FnIII-1/-2 domains and α-CT helix of IGF1R. In contrast, the VHH-IR5 sdAb-mediated changes in conformational dynamics were limited to the α-CT helix and its immediate vicinity (L1 domain). High-resolution NMR spectroscopy titration data and linear peptide scanning demonstrated that VHH-IR5 has high-affinity binding interactions with a peptide sequence around the C-terminal region of the α-CT helix. Taken together, these results define a core linear epitope for VHH-IR5 within the α-CT helix, overlapping the IGF-1 binding site, and suggest a potential role for the α-CT helix in sdAb-mediated transcytosis.
Conflict of interest statement
The authors declare no competing interests.
Figures
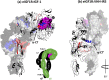

Similar articles
-
Epitope mapping of a blood-brain barrier crossing antibody targeting the cysteine-rich region of IGF1R using hydrogen-exchange mass spectrometry enabled by electrochemical reduction.J Biochem. 2023 Feb 3;173(2):95-105. doi: 10.1093/jb/mvac088. J Biochem. 2023. PMID: 36346120
-
Epitope-specific mechanisms of IGF1R inhibition by ganitumab.PLoS One. 2013;8(2):e55135. doi: 10.1371/journal.pone.0055135. Epub 2013 Feb 1. PLoS One. 2013. PMID: 23383308 Free PMC article.
-
Targeting insulin-like growth factor-1 receptor (IGF1R) for brain delivery of biologics.FASEB J. 2022 Mar;36(3):e22208. doi: 10.1096/fj.202101644R. FASEB J. 2022. PMID: 35192204
-
[The structural organization of binding determinants in insulin-like growth factor-I (IGF-I) molecule].Zh Evol Biokhim Fiziol. 2010 Jan-Feb;46(1):74-94. Zh Evol Biokhim Fiziol. 2010. PMID: 20297673 Review. Russian.
-
Single domain antibody-based vectors in the delivery of biologics across the blood-brain barrier: a review.Drug Deliv Transl Res. 2021 Oct;11(5):1818-1828. doi: 10.1007/s13346-020-00873-7. Epub 2020 Nov 5. Drug Deliv Transl Res. 2021. PMID: 33155179 Review.
Cited by
-
Nanobody engineering: computational modelling and design for biomedical and therapeutic applications.FEBS Open Bio. 2025 Feb;15(2):236-253. doi: 10.1002/2211-5463.13850. Epub 2024 Jun 19. FEBS Open Bio. 2025. PMID: 38898362 Free PMC article. Review.
-
Leveraging neonatal Fc receptor (FcRn) to enhance antibody transport across the blood brain barrier.Nat Commun. 2025 May 3;16(1):4143. doi: 10.1038/s41467-025-59447-1. Nat Commun. 2025. PMID: 40319060 Free PMC article.
-
In vivo brain delivery of BBB-enabled iduronate 2-sulfatase in rats.Fluids Barriers CNS. 2025 Jan 14;22(1):7. doi: 10.1186/s12987-024-00617-6. Fluids Barriers CNS. 2025. PMID: 39810248 Free PMC article.
-
Neuroprotective Effect of Wharton's Jelly-Derived Mesenchymal Stem Cell-Conditioned Medium (WJMSC-CM) on Diabetes-Associated Cognitive Impairment by Improving Oxidative Stress, Neuroinflammation, and Apoptosis.Stem Cells Int. 2023 Apr 11;2023:7852394. doi: 10.1155/2023/7852394. eCollection 2023. Stem Cells Int. 2023. PMID: 37081849 Free PMC article.
-
Brain Delivery of IGF1R5, a Single-Domain Antibody Targeting Insulin-like Growth Factor-1 Receptor.Pharmaceutics. 2022 Jul 12;14(7):1452. doi: 10.3390/pharmaceutics14071452. Pharmaceutics. 2022. PMID: 35890347 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

