Effects of maoto (ma-huang-tang) on host lipid mediator and transcriptome signature in influenza virus infection
- PMID: 33608574
- PMCID: PMC7896050
- DOI: 10.1038/s41598-021-82707-1
Effects of maoto (ma-huang-tang) on host lipid mediator and transcriptome signature in influenza virus infection
Abstract
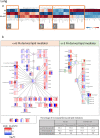
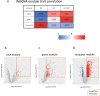
Maoto, a traditional kampo medicine, has been clinically prescribed for influenza infection and is reported to relieve symptoms and tissue damage. In this study, we evaluated the effects of maoto as an herbal multi-compound medicine on host responses in a mouse model of influenza infection. On the fifth day of oral administration to mice intranasally infected with influenza virus [A/PR/8/34 (H1N1)], maoto significantly improved survival rate, decreased viral titer, and ameliorated the infection-induced phenotype as compared with control mice. Analysis of the lung and plasma transcriptome and lipid mediator metabolite profile showed that maoto altered the profile of lipid mediators derived from ω-6 and ω-3 fatty acids to restore a normal state, and significantly up-regulated the expression of macrophage- and T-cell-related genes. Collectively, these results suggest that maoto regulates the host's inflammatory response by altering the lipid mediator profile and thereby ameliorating the symptoms of influenza.
Conflict of interest statement
A. N., N.K., C.S., K. Oh., S. I., A.S., K. Og. and M. Y. are employed by Tsumura & Co. H. Ku., S. N., A. Y., Y. M., and H. Ki. have financial interests in Tsumura & Co. relevant to this research. The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

