Prostate adenocarcinoma and COVID-19: The possible impacts of TMPRSS2 expressions in susceptibility to SARS-CoV-2
- PMID: 33609069
- PMCID: PMC8013364
- DOI: 10.1111/jcmm.16385
Prostate adenocarcinoma and COVID-19: The possible impacts of TMPRSS2 expressions in susceptibility to SARS-CoV-2
Abstract
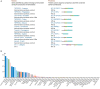
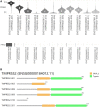
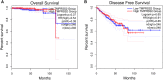
TMPRSS2 (OMIM: 602060) is a cellular protease involved in many physiological and pathological processes, and it facilitates entry of viruses such as SARS-CoV-2 into host cells. It is important to predict the prostate's susceptibility to SARS-CoV-2 infection in cancer patients and the disease outcome by assessing TMPRSS2 expression in cancer tissues. In this study, we conducted the expression profiles of the TMPRSS2 gene for COVID-19 in different normal tissues and PRAD (prostate adenocarcinoma) tumour tissues. TMPRSS2 is highly expressed in normal tissues including the small intestine, prostate, pancreas, salivary gland, colon, stomach, seminal vesicle and lung, and is increased in PRAD tissues, indicating that SARS-CoV-2 might attack not only the lungs and other normal organs, but also in PRAD cancer tissues. Hypomethylation of TMPRSS2 promoter may not be the mechanism for TMPRSS2 overexpression in PRAD tissues and PRAD pathogenesis. TMPRSS2 expresses eleven isoforms in PRAD tissues, with the TMPRSS2-001 isoform expressed highest and followed by TMPRSS2-201. Further isoform structures prediction showed that these two highly expressed isoforms have both SRCR_2 and Trypsin (Tryp_SPc) domains, which may be essential for TMPRSS2 functional roles for tumorigenesis and entry for SARS-CoV-2 in PRAD patients. Analyses of functional annotation and enrichment in TMPRSS2 showed that TMPRSS2 is mostly enriched in regulation of viral entry into host cells, protein processing and serine-type peptidase activity. TMPRSS2 is also associated with prostate gland cancer cell expression, different complex(es) formation, human influenza and carcinoma, pathways in prostate cancer, influenza A, and transcriptional misregulation in cancer. Altogether, even though high expression of TMPRSS2 may not be favourable for PRAD patient's survival, increased expression in these patients should play roles in susceptibility of the SARS-CoV-2 infection and clinical severity for COVID-19, highlighting the value of protective actions of PRAD cases by targeting or androgen-mediated therapeutic strategies in the COVID-19 pandemic.
Keywords: COVID-19; SARS-CoV-2; TMPRSS2 gene; prostate adenocarcinoma; susceptibility.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
None.
Figures


References
-
- Paoloni‐Giacobino A, Chen H, Peitsch MC, Rossier C, Antonarakis SE. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics. 1997;44:309‐320. - PubMed
-
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644‐648. - PubMed
-
- Ko CJ, Huang CC, Lin HY, et al. Androgen‐induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Can Res. 2015;75:2949‐2960. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 00031726/Special Training Program for Young Science and Technology Talents from Southwest Medical University
- 2018LZXNYD-YL01/The Joint Research Foundation of Luzhou City and Southwest Medical University
- 81672887/National Natural Science Foundation of China
- 82073263/National Natural Science Foundation of China
- 30371493/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

