Strong functional data for pathogenicity or neutrality classify BRCA2 DNA-binding-domain variants of uncertain significance
- PMID: 33609447
- PMCID: PMC8008494
- DOI: 10.1016/j.ajhg.2021.02.005
Strong functional data for pathogenicity or neutrality classify BRCA2 DNA-binding-domain variants of uncertain significance
Abstract
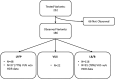
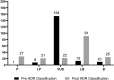
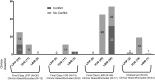
Determination of the clinical relevance of rare germline variants of uncertain significance (VUSs) in the BRCA2 cancer predisposition gene remains a challenge as a result of limited availability of data for use in classification models. However, laboratory-based functional data derived from validated functional assays of known sensitivity and specificity may influence the interpretation of VUSs. We evaluated 252 missense VUSs from the BRCA2 DNA-binding domain by using a homology-directed DNA repair (HDR) assay and identified 90 as non-functional and 162 as functional. The functional assay results were integrated with other available data sources into an ACMG/AMP rules-based classification framework used by a hereditary cancer testing laboratory. Of the 186 missense variants observed by the testing laboratory, 154 were classified as VUSs without functional data. However, after applying protein functional data, 86% (132/154) of the VUSs were reclassified as either likely pathogenic/pathogenic (39/132) or likely benign/benign (93/132), which impacted testing results for 1,900 individuals. These results indicate that validated functional assay data can have a substantial impact on VUS classification and associated clinical management for many individuals with inherited alterations in BRCA2.
Keywords: ACMG/AMP; BRCA2; breast cancer; functional assay; predisposition gene; variant of uncertain significance.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
K.F., K.M.D., A.M.D., H.L., M.E.R., E.C., and T.P. are employees of Ambry Genetics. F.J.C. has received consulting fees from Astrazeneca. The remaining authors declare no competing interests.
Figures
References
-
- Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.A., Mooij T.M., Roos-Blom M.J., Jervis S., van Leeuwen F.E., Milne R.L., Andrieu N., BRCA1 and BRCA2 Cohort Consortium Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317:2402–2416. - PubMed
-
- Kim G., Ison G., McKee A.E., Zhang H., Tang S., Gwise T., Sridhara R., Lee E., Tzou A., Philip R. FDA Approval Summary: Olaparib Monotherapy in Patients with Deleterious Germline BRCA-Mutated Advanced Ovarian Cancer Treated with Three or More Lines of Chemotherapy. Clin. Cancer Res. 2015;21:4257–4261. - PubMed
-
- Balasubramaniam S., Beaver J.A., Horton S., Fernandes L.L., Tang S., Horne H.N., Liu J., Liu C., Schrieber S.J., Yu J. FDA Approval Summary: Rucaparib for the Treatment of Patients with Deleterious BRCA Mutation-Associated Advanced Ovarian Cancer. Clin. Cancer Res. 2017;23:7165–7170. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

