Effects of H2A.B incorporation on nucleosome structures and dynamics
- PMID: 33609493
- PMCID: PMC8105712
- DOI: 10.1016/j.bpj.2021.01.036
Effects of H2A.B incorporation on nucleosome structures and dynamics
Abstract
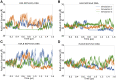
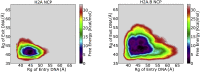
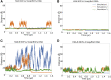
The H2A.B histone variant is an epigenetic regulator involved in transcriptional upregulation, DNA synthesis, and splicing that functions by replacing the canonical H2A histone in the nucleosome core particle. Introduction of H2A.B results in less compact nucleosome states with increased DNA unwinding and accessibility at the nucleosomal entry and exit sites. Despite being well characterized experimentally, the molecular mechanisms by which H2A.B incorporation alters nucleosome stability and dynamics remain poorly understood. To study the molecular mechanisms of H2A.B, we have performed a series of conventional and enhanced sampling molecular dynamics simulation of H2A.B- and canonical H2A-containing nucleosomes. Results of conventional simulations show that H2A.B weakens protein-protein and protein-DNA interactions at specific locations throughout the nucleosome. These weakened interactions result in significantly more DNA opening from both the entry and exit sites in enhanced sampling simulations. Furthermore, free energy profiles show that H2A.B-containing nucleosomes have significantly broader free wells and that H2A.B allows for sampling of states with increased DNA breathing, which are shown to be stable on the hundreds of nanoseconds timescale with further conventional simulations. Together, our results show the molecular mechanisms by which H2A.B creates less compacted nucleosome states as a means of increasing genetic accessibility and gene transcription.
Copyright © 2021 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Nucleosomal DNA unwinding pathway through canonical and non-canonical histone disassembly.Commun Biol. 2024 Sep 14;7(1):1144. doi: 10.1038/s42003-024-06856-5. Commun Biol. 2024. PMID: 39277674 Free PMC article.
-
Unique Dynamics in Asymmetric macroH2A-H2A Hybrid Nucleosomes Result in Increased Complex Stability.J Phys Chem B. 2019 Jan 17;123(2):419-427. doi: 10.1021/acs.jpcb.8b10668. Epub 2019 Jan 8. J Phys Chem B. 2019. PMID: 30557018 Free PMC article.
-
Effects of MacroH2A and H2A.Z on Nucleosome Dynamics as Elucidated by Molecular Dynamics Simulations.Biophys J. 2016 Jan 19;110(2):327-337. doi: 10.1016/j.bpj.2015.12.015. Biophys J. 2016. PMID: 26789756 Free PMC article.
-
Histone H2A variants in nucleosomes and chromatin: more or less stable?Nucleic Acids Res. 2012 Nov;40(21):10719-41. doi: 10.1093/nar/gks865. Epub 2012 Sep 21. Nucleic Acids Res. 2012. PMID: 23002134 Free PMC article. Review.
-
Reconciling the positive and negative roles of histone H2A.Z in gene transcription.Epigenetics. 2010 May 16;5(4):267-72. doi: 10.4161/epi.5.4.11520. Epub 2010 May 16. Epigenetics. 2010. PMID: 20364108 Review.
Cited by
-
Nuclear envelope, chromatin organizers, histones, and DNA: The many achilles heels exploited across cancers.Front Cell Dev Biol. 2022 Dec 16;10:1068347. doi: 10.3389/fcell.2022.1068347. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36589746 Free PMC article. Review.
-
Always on the Move: Overview on Chromatin Dynamics within Nuclear Processes.Biochemistry. 2025 May 20;64(10):2138-2153. doi: 10.1021/acs.biochem.5c00114. Epub 2025 May 1. Biochemistry. 2025. PMID: 40312022 Free PMC article. Review.
-
Casting histone variants during mammalian reproduction.Chromosoma. 2023 Sep;132(3):153-165. doi: 10.1007/s00412-023-00803-9. Epub 2023 Jun 22. Chromosoma. 2023. PMID: 37347315 Free PMC article. Review.
-
Nucleosomal DNA unwinding pathway through canonical and non-canonical histone disassembly.Commun Biol. 2024 Sep 14;7(1):1144. doi: 10.1038/s42003-024-06856-5. Commun Biol. 2024. PMID: 39277674 Free PMC article.
-
The effects of RNA.DNA-DNA triple helices on nucleosome structures and dynamics.Biophys J. 2023 Apr 4;122(7):1229-1239. doi: 10.1016/j.bpj.2023.02.013. Epub 2023 Feb 16. Biophys J. 2023. PMID: 36798026 Free PMC article.
References
-
- Kornberg R.D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. - PubMed
-
- Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. - PubMed
-
- Olins A.L., Olins D.E. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

