Hybrid ensemble model for differential diagnosis between COVID-19 and common viral pneumonia by chest X-ray radiograph
- PMID: 33610001
- PMCID: PMC7966819
- DOI: 10.1016/j.compbiomed.2021.104252
Hybrid ensemble model for differential diagnosis between COVID-19 and common viral pneumonia by chest X-ray radiograph
Abstract
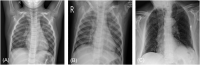
Background: Chest X-ray radiography (CXR) has been widely considered as an accessible, feasible, and convenient method to evaluate suspected patients' lung involvement during the COVID-19 pandemic. However, with the escalating number of suspected cases, traditional diagnosis via CXR fails to deliver results within a short period of time. Therefore, it is crucial to employ artificial intelligence (AI) to enhance CXRs for obtaining quick and accurate diagnoses. Previous studies have reported the feasibility of utilizing deep learning methods to screen for COVID-19 using CXR and CT results. However, these models only use a single deep learning network for chest radiograph detection; the accuracy of this approach required further improvement.
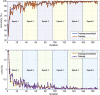
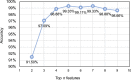
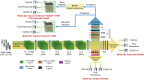
Methods: In this study, we propose a three-step hybrid ensemble model, including a feature extractor, a feature selector, and a classifier. First, a pre-trained AlexNet with an improved structure extracts the original image features. Then, the ReliefF algorithm is adopted to sort the extracted features, and a trial-and-error approach is used to select the n most important features to reduce the feature dimension. Finally, an SVM classifier provides classification results based on the n selected features.
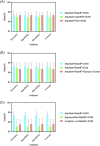
Results: Compared to five existing models (InceptionV3: 97.916 ± 0.408%; SqueezeNet: 97.189 ± 0.526%; VGG19: 96.520 ± 1.220%; ResNet50: 97.476 ± 0.513%; ResNet101: 98.241 ± 0.209%), the proposed model demonstrated the best performance in terms of overall accuracy rate (98.642 ± 0.398%). Additionally, compared to the existing models, the proposed model demonstrates a considerable improvement in classification time efficiency (SqueezeNet: 6.602 ± 0.001s; InceptionV3: 12.376 ± 0.002s; ResNet50: 10.952 ± 0.001s; ResNet101: 18.040 ± 0.002s; VGG19: 16.632 ± 0.002s; proposed model: 5.917 ± 0.001s).
Conclusion: The model proposed in this article is practical and effective, and can provide high-precision COVID-19 CXR detection. We demonstrated its suitability to aid medical professionals in distinguishing normal CXRs, viral pneumonia CXRs and COVID-19 CXRs efficiently on small sample sizes.
Keywords: COVID-19; Dimension reduction; Transfer learning; X-ray imaging.
Copyright © 2021. Published by Elsevier Ltd.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures




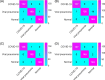
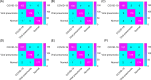

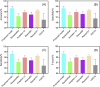
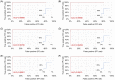

Similar articles
-
Computer aid screening of COVID-19 using X-ray and CT scan images: An inner comparison.J Xray Sci Technol. 2021;29(2):197-210. doi: 10.3233/XST-200784. J Xray Sci Technol. 2021. PMID: 33492267
-
MCSC-Net: COVID-19 detection using deep-Q-neural network classification with RFNN-based hybrid whale optimization.J Xray Sci Technol. 2023;31(3):483-509. doi: 10.3233/XST-221360. J Xray Sci Technol. 2023. PMID: 36872839
-
A Deep Learning Model for Diagnosing COVID-19 and Pneumonia through X-ray.Curr Med Imaging. 2023;19(4):333-346. doi: 10.2174/1573405618666220610093740. Curr Med Imaging. 2023. PMID: 35692156
-
COVID-19 diagnosis: A comprehensive review of pre-trained deep learning models based on feature extraction algorithm.Results Eng. 2023 Jun;18:101020. doi: 10.1016/j.rineng.2023.101020. Epub 2023 Mar 16. Results Eng. 2023. PMID: 36945336 Free PMC article. Review.
-
Thoracic imaging tests for the diagnosis of COVID-19.Cochrane Database Syst Rev. 2020 Sep 30;9:CD013639. doi: 10.1002/14651858.CD013639.pub2. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2020 Nov 26;11:CD013639. doi: 10.1002/14651858.CD013639.pub3. PMID: 32997361 Updated.
Cited by
-
A brief review and scientometric analysis on ensemble learning methods for handling COVID-19.Heliyon. 2024 Feb 20;10(4):e26694. doi: 10.1016/j.heliyon.2024.e26694. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38420425 Free PMC article.
-
A stacked ensemble for the detection of COVID-19 with high recall and accuracy.Comput Biol Med. 2021 Aug;135:104608. doi: 10.1016/j.compbiomed.2021.104608. Epub 2021 Jun 30. Comput Biol Med. 2021. PMID: 34247135 Free PMC article.
-
Medical imaging and computational image analysis in COVID-19 diagnosis: A review.Comput Biol Med. 2021 Aug;135:104605. doi: 10.1016/j.compbiomed.2021.104605. Epub 2021 Jun 23. Comput Biol Med. 2021. PMID: 34175533 Free PMC article. Review.
-
Ensemble learning for multi-class COVID-19 detection from big data.PLoS One. 2023 Oct 11;18(10):e0292587. doi: 10.1371/journal.pone.0292587. eCollection 2023. PLoS One. 2023. Retraction in: PLoS One. 2025 Apr 7;20(4):e0321416. doi: 10.1371/journal.pone.0321416. PMID: 37819992 Free PMC article. Retracted.
-
Explainable artificial intelligence-based edge fuzzy images for COVID-19 detection and identification.Appl Soft Comput. 2022 Jul;123:108966. doi: 10.1016/j.asoc.2022.108966. Epub 2022 May 13. Appl Soft Comput. 2022. PMID: 35582662 Free PMC article.
References
-
- Radiology U.C.L.A. COVID-19 chest X-ray guideline. https://www.uclahealth.org/radiology/covid-19-chest-x-ray-guideline
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

