Structural characterization of a PCP-R didomain from an archaeal nonribosomal peptide synthetase reveals novel interdomain interactions
- PMID: 33610550
- PMCID: PMC8024701
- DOI: 10.1016/j.jbc.2021.100432
Structural characterization of a PCP-R didomain from an archaeal nonribosomal peptide synthetase reveals novel interdomain interactions
Abstract
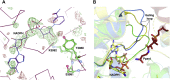
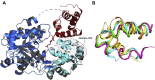
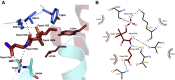
Nonribosomal peptide synthetases (NRPSs) are multimodular enzymes that produce a wide range of bioactive peptides, such as siderophores, toxins, and antibacterial and insecticidal agents. NRPSs are dynamic proteins characterized by extensive interdomain communications as a consequence of their assembly-line mode of synthesis. Hence, crystal structures of multidomain fragments of NRPSs have aided in elucidating crucial interdomain interactions that occur during different steps of the NRPS catalytic cycle. One crucial yet unexplored interaction is that between the reductase (R) domain and the peptide carrier protein (PCP) domain. R domains are members of the short-chain dehydrogenase/reductase family and function as termination domains that catalyze the reductive release of the final peptide product from the terminal PCP domain of the NRPS. Here, we report the crystal structure of an archaeal NRPS PCP-R didomain construct. This is the first NRPS R domain structure to be determined together with the upstream PCP domain and is also the first structure of an archaeal NRPS to be reported. The structure reveals that a novel helix-turn-helix motif, found in NRPS R domains but not in other short-chain dehydrogenase/reductase family members, plays a major role in the interface between the PCP and R domains. The information derived from the described PCP-R interface will aid in gaining further mechanistic insights into the peptide termination reaction catalyzed by the R domain and may have implications in engineering NRPSs to synthesize novel peptide products.
Keywords: X-ray crystallography; archaea; nonribosomal peptide synthetase; peptide biosynthesis; peptide carrier protein domain; protein–protein interaction; reductase domain; short-chain dehydrogenase/reductase; structure–function.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Marahiel M.A., Stachelhaus T., Mootz H.D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 1997;97:2651–2673. - PubMed
-
- Süssmuth R.D., Mainz A. Nonribosomal peptide synthesis—principles and prospects. Angew. Chem. Int. Edition. 2017;56:3770–3821. - PubMed
-
- Du L., Lou L. PKS and NRPS release mechanisms. Nat. Product Rep. 2010;27:255–278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

