Hsa_circ_0038383-mediated competitive endogenous RNA network in recurrent implantation failure
- PMID: 33611311
- PMCID: PMC7950293
- DOI: 10.18632/aging.202590
Hsa_circ_0038383-mediated competitive endogenous RNA network in recurrent implantation failure
Abstract
Background: Inadequate endometrial receptivity contributes to recurrent implantation failure (RIF) during IVF-embryo transfer. Though multiple circRNAs have been confirmed differentially expression in RIF, the potential function of novel circRNAs needed to be detected.
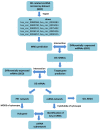
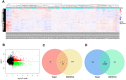
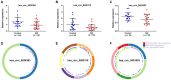
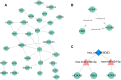
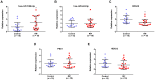
Results: The top ten DEcircRNAs were selected as initial candidates. A ceRNA network was conducted on the basis of circRNA-miRNA-mRNA potential interaction, consisting of 10 DEcircRNAs, 28 DEmiRNAs and 59 DEmRNAs. Three down-regulation circRNAs with high degree of connectivity were verified by RT-qPCR, and results suggested that only hsa_circ_0038383 was significantly downregulation in RIF compared with control group. Subsequently, three hub genes (HOXA3, HOXA9 and PBX1) were identified as hub genes. Ultimately, a subnetwork was determined based on one DEcircRNA (hsa_circ_0038383), two DEmiRNAs (has-miR-196b-5p and has-miR-424-5p), and three DEmRNAs (HOXA3, HOXA9 and PBX1). Following verification, hsa_circ_0038383/miR-196b-5p/HOXA9 axis may be a key pathway in affecting RIF.
Conclusion: In summary, a hsa_circ_0038383-mediated ceRNA network related to RIF was proposed. This network provided new insight into exploring potential biomarkers for diagnosis and clinical treatment of RIF.
Methods: We retrieved the expression profiles of RIF from GEO databases (circRNA, microRNA and mRNA) and constructed a competing endogenous RNAs (ceRNA) network based on predicted circRNA-miRNA and miRNA-mRNA pairs. The expression levels of three hub DEcircRNAs identified by cytoscape were validated by RT-qPCR.
Keywords: ceRNA; circRNA; endometrial receptivity; miRNA; recurrent implantation failure.
Conflict of interest statement
Figures




Similar articles
-
Construction and Bioinformatics Analysis of circRNA-miRNA-mRNA Network in Acute Myocardial Infarction.Front Genet. 2022 Mar 29;13:854993. doi: 10.3389/fgene.2022.854993. eCollection 2022. Front Genet. 2022. PMID: 35422846 Free PMC article.
-
Construction of a circRNA-miRNA-mRNA Regulatory Network for Coronary Artery Disease by Bioinformatics Analysis.Cardiol Res Pract. 2022 Feb 16;2022:4017082. doi: 10.1155/2022/4017082. eCollection 2022. Cardiol Res Pract. 2022. PMID: 35223093 Free PMC article.
-
Comprehensive analysis of circRNA-miRNA-mRNA network related to angiogenesis in recurrent implantation failure.BMC Med Genomics. 2024 Jul 30;17(1):193. doi: 10.1186/s12920-024-01944-1. BMC Med Genomics. 2024. PMID: 39080700 Free PMC article.
-
RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection.Front Cell Infect Microbiol. 2020 Mar 13;10:97. doi: 10.3389/fcimb.2020.00097. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32232013 Free PMC article. Review.
-
MicroRNA Signatures in Endometrial Receptivity-Unlocking Their Role in Embryo Implantation and IVF Success: A Systematic Review.Biomedicines. 2025 May 13;13(5):1189. doi: 10.3390/biomedicines13051189. Biomedicines. 2025. PMID: 40427016 Free PMC article. Review.
Cited by
-
hsa_circ_0004121 and hsa_circ_0030162 Differentially Expressed in Plasma of Patients with Recurrent Implantation Failure.Rep Biochem Mol Biol. 2024 Oct;13(3):428-437. doi: 10.61186/rbmb.13.3.428. Rep Biochem Mol Biol. 2024. PMID: 40330557 Free PMC article.
-
Construction of circRNA-miRNA-mRNA network in the pathogenesis of recurrent implantation failure using integrated bioinformatics study.J Cell Mol Med. 2022 Mar;26(6):1853-1864. doi: 10.1111/jcmm.16586. Epub 2021 May 7. J Cell Mol Med. 2022. PMID: 33960101 Free PMC article.
-
Upregulated RPA2 in endometrial tissues of repeated implantation failure patients impairs the endometrial decidualization.J Assist Reprod Genet. 2023 Nov;40(11):2739-2750. doi: 10.1007/s10815-023-02946-1. Epub 2023 Oct 13. J Assist Reprod Genet. 2023. PMID: 37831348 Free PMC article.
-
Identification of a circRNA/miRNA/mRNA ceRNA Network as a Cell Cycle-Related Regulator for Chronic Sinusitis with Nasal Polyps.J Inflamm Res. 2022 Apr 23;15:2601-2615. doi: 10.2147/JIR.S358387. eCollection 2022. J Inflamm Res. 2022. PMID: 35494315 Free PMC article.
-
Construction and analysis of a survival-associated competing endogenous RNA network in breast cancer.Front Surg. 2023 Jan 6;9:1021195. doi: 10.3389/fsurg.2022.1021195. eCollection 2022. Front Surg. 2023. PMID: 36684328 Free PMC article.
References
-
- Kupka MS, D'Hooghe T, Ferraretti AP, de Mouzon J, Erb K, Castilla JA, Calhaz-Jorge C, De Geyter Ch, Goossens V, and European IVF-Monitoring Consortium (EIM), and European Society of Human Reproduction and Embryology (ESHRE). Assisted reproductive technology in Europe, 2011: results generated from European registers by ESHRE. Hum Reprod. 2016; 31:233–48. 10.1093/humrep/dev319 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

