Proteogenomic Insights into the Physiology of Marine, Sulfate-Reducing, Filamentous Desulfonema limicola and Desulfonema magnum
- PMID: 33611323
- PMCID: PMC8315694
- DOI: 10.1159/000513383
Proteogenomic Insights into the Physiology of Marine, Sulfate-Reducing, Filamentous Desulfonema limicola and Desulfonema magnum
Abstract
The genus Desulfonema belongs to the deltaproteobacterial family Desulfobacteraceae and comprises marine, sulfate-reducing bacteria that form filaments and move by gliding. This study reports on the complete, manually annotated genomes of Dn. limicola 5ac10T (6.91 Mbp; 6,207 CDS) and Dn. magnum 4be13T (8.03 Mbp; 9,970 CDS), integrated with substrate-specific proteome profiles (8 vs. 11). The richness in mobile genetic elements is shared with other Desulfobacteraceae members, corroborating horizontal gene transfer as major driver in shaping the genomes of this family. The catabolic networks of Dn. limicola and Dn. magnum have the following general characteristics: 98 versus 145 genes assigned (having genomic shares of 1.7 vs. 2.2%), 92.5 versus 89.7% proteomic coverage, and scattered gene clusters for substrate degradation and energy metabolism. The Dn. magnum typifying capacity for aromatic compound degradation (e.g., p-cresol, 3-phenylpropionate) requires 48 genes organized in operon-like structures (87.7% proteomic coverage; no homologs in Dn. limicola). The protein complements for aliphatic compound degradation, central pathways, and energy metabolism are highly similar between both genomes and were identified to a large extent (69-96%). The differential protein profiles revealed a high degree of substrate-specificity for peripheral reaction sequences (forming central intermediates), agreeing with the high number of sensory/regulatory proteins predicted for both strains. By contrast, central pathways and modules of the energy metabolism were constitutively formed under the tested substrate conditions. In accord with their natural habitats that are subject to fluctuating changes of physicochemical parameters, both Desulfonema strains are well equipped to cope with various stress conditions. Next to superoxide dismutase and catalase also desulfoferredoxin and rubredoxin oxidoreductase are formed to counter exposure to molecular oxygen. A variety of proteases and chaperones were detected that function in maintaining cellular homeostasis upon heat or cold shock. Furthermore, glycine betaine/proline betaine transport systems can respond to hyperosmotic stress. Gliding movement probably relies on twitching motility via type-IV pili or adventurous motility. Taken together, this proteogenomic study demonstrates the adaptability of Dn. limicola and Dn. magnum to its dynamic habitats by means of flexible catabolism and extensive stress response capacities.
Keywords: Anaerobic degradation; Aromatic compounds; Complete genome; Desulfonema limicola; Desulfonema magnum; Differential proteomics; Metabolism; Physiology; Stress response; Sulfate reduction.
© 2021 The Author(s) Published by S. Karger AG, Basel.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



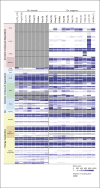
Similar articles
-
Physiology, phylogenetic relationships, and ecology of filamentous sulfate-reducing bacteria (genus desulfonema).Arch Microbiol. 1999 Oct;172(4):193-203. doi: 10.1007/s002030050760. Arch Microbiol. 1999. PMID: 10525735
-
Genome and catabolic subproteomes of the marine, nutritionally versatile, sulfate-reducing bacterium Desulfococcus multivorans DSM 2059.BMC Genomics. 2016 Nov 15;17(1):918. doi: 10.1186/s12864-016-3236-7. BMC Genomics. 2016. PMID: 27846794 Free PMC article.
-
Complete genome, catabolic sub-proteomes and key-metabolites of Desulfobacula toluolica Tol2, a marine, aromatic compound-degrading, sulfate-reducing bacterium.Environ Microbiol. 2013 May;15(5):1334-55. doi: 10.1111/j.1462-2920.2012.02885.x. Epub 2012 Oct 23. Environ Microbiol. 2013. PMID: 23088741
-
The Catabolic Network of Aromatoleum aromaticum EbN1T.Microb Physiol. 2024;34(1):1-77. doi: 10.1159/000534425. Epub 2023 Oct 10. Microb Physiol. 2024. PMID: 37816339 Review.
-
Gliding motility in bacteria: insights from studies of Myxococcus xanthus.Microbiol Mol Biol Rev. 1999 Sep;63(3):621-41. doi: 10.1128/MMBR.63.3.621-641.1999. Microbiol Mol Biol Rev. 1999. PMID: 10477310 Free PMC article. Review.
Cited by
-
Indications for a genetic basis for big bacteria and description of the giant cable bacterium Candidatus Electrothrix gigas sp. nov.Microbiol Spectr. 2023 Sep 21;11(5):e0053823. doi: 10.1128/spectrum.00538-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37732806 Free PMC article.
-
Key role of Desulfobacteraceae in C/S cycles of marine sediments is based on congeneric catabolic-regulatory networks.Sci Adv. 2025 Mar 7;11(10):eads5631. doi: 10.1126/sciadv.ads5631. Epub 2025 Mar 7. Sci Adv. 2025. PMID: 40053579 Free PMC article.
-
Large Filamentous Bacteria Isolated From Sulphidic Sediments Reveal Novel Species and Distinct Energy and Defence Mechanisms for Survival.Environ Microbiol. 2025 Mar;27(3):e70083. doi: 10.1111/1462-2920.70083. Environ Microbiol. 2025. PMID: 40103259 Free PMC article.
-
Hidden Markov Model-Based Prokaryotic Genome Space Mining Reveals the Widespread Pervasiveness of Complex I and Its Potential Evolutionary Scheme.Genome Biol Evol. 2025 Jul 30;17(8):evaf154. doi: 10.1093/gbe/evaf154. Genome Biol Evol. 2025. PMID: 40795046 Free PMC article.
-
RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.Science. 2022 Nov 25;378(6622):882-889. doi: 10.1126/science.add7347. Epub 2022 Nov 3. Science. 2022. PMID: 36423304 Free PMC article.
References
-
- Aeckersberg F, Bak F, Widdel F. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch Microbiol. 1991 Jul;156((1)):5–14.
-
- Amann J, Lange D, Schüler M, Rabus R. Substrate-dependent regulation of carbon catabolism in marine sulfate-reducing Desulfobacterium autotrophicum HRM2. J Mol Microbiol Biotechnol. 2010 Apr;18((2)):74–84. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

