Full-Length 16S rRNA and ITS Gene Sequencing Revealed Rich Microbial Flora in Roots of Cycas spp. in China
- PMID: 33613025
- PMCID: PMC7868495
- DOI: 10.1177/1176934321989713
Full-Length 16S rRNA and ITS Gene Sequencing Revealed Rich Microbial Flora in Roots of Cycas spp. in China
Abstract
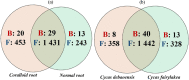
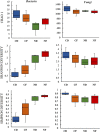
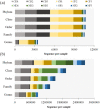
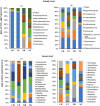
Cycads have developed a complex root system categorized either as normal or coralloid roots. Past literatures revealed that a great diversity of key microbes is associated with these roots. This recent study aims to comprehensively determine the diversity and community structure of bacteria and fungi associated with the roots of two Cycas spp. endemic to China, Cycas debaoensis Zhong & Chen and Cycas fairylakea D.Y. Wang using high-throughput amplicon sequencing of the full-length 16S rRNA (V1-V9 hypervariable) and short fragment ITS region. The total DNA from 12 root samples were extracted, amplified, sequenced, and analyzed. Resulting sequences were clustered into 61 bacteria and 2128 fungal OTUs. Analysis of community structure revealed that the coralloid roots were dominated mostly by the nitrogen-fixer Nostocaceae but also contain other non-diazotrophic bacteria. The sequencing of entire 16S rRNA gene identified four different strains of cyanobacteria under the heterocystous genera Nostoc and Desmonostoc. Meanwhile, the top bacterial families in normal roots were Xanthobacteraceae, Burkholderiaceae, and Bacillaceae. Moreover, a diverse fungal community was also found in the roots of cycads and the predominating families were Ophiocordycipitaceae, Nectriaceae, Bionectriaceae, and Trichocomaceae. Our results demonstrated that bacterial diversity in normal roots of C. fairylakea is higher in richness and abundance than C. debaoensis. On the other hand, a slight difference, albeit insignificant, was noted for the diversity of fungi among root types and host species as the number of shared taxa is relatively high (67%). Our results suggested that diverse microbes are present in roots of cycads which potentially interact together to support cycads survival. Our study provided additional knowledge on the microbial diversity and composition in cycads and thus expanding our current knowledge on cycad-microbe association. Our study also considered the possible impact of ex situ conservation on cyanobiont community of cycads.
Keywords: Cycas; coralloid root; diversity; metagenomics; root microbiome.
© The Author(s) 2021.
Conflict of interest statement
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures


Similar articles
-
Core Endophytic Bacteria and Their Roles in the Coralloid Roots of Cultivated Cycas revoluta (Cycadaceae).Microorganisms. 2023 Sep 21;11(9):2364. doi: 10.3390/microorganisms11092364. Microorganisms. 2023. PMID: 37764208 Free PMC article.
-
Core Microbiome and Microbial Community Structure in Coralloid Roots of Cycas in Ex Situ Collection of Kunming Botanical Garden in China.Microorganisms. 2023 Aug 24;11(9):2144. doi: 10.3390/microorganisms11092144. Microorganisms. 2023. PMID: 37763988 Free PMC article.
-
Highly diverse endophytes in roots of Cycas bifida (Cycadaceae), an ancient but endangered gymnosperm.J Microbiol. 2018 May;56(5):337-345. doi: 10.1007/s12275-018-7438-3. Epub 2018 May 2. J Microbiol. 2018. PMID: 29721831
-
Perspectives on Endosymbiosis in Coralloid Roots: Association of Cycads and Cyanobacteria.Front Microbiol. 2019 Aug 14;10:1888. doi: 10.3389/fmicb.2019.01888. eCollection 2019. Front Microbiol. 2019. PMID: 31474965 Free PMC article. Review.
-
Structure and Biosynthesis of Desmamides A-C, Lipoglycopeptides from the Endophytic Cyanobacterium Desmonostoc muscorum LEGE 12446.J Nat Prod. 2022 Jul 22;85(7):1704-1714. doi: 10.1021/acs.jnatprod.2c00162. Epub 2022 Jul 6. J Nat Prod. 2022. PMID: 35793792 Free PMC article. Review.
Cited by
-
Possible role of arbuscular mycorrhizal fungi and associated bacteria in the recruitment of endophytic bacterial communities by plant roots.Mycorrhiza. 2021 Oct;31(5):527-544. doi: 10.1007/s00572-021-01040-7. Epub 2021 Jul 20. Mycorrhiza. 2021. PMID: 34286366 Free PMC article. Review.
-
Core Endophytic Bacteria and Their Roles in the Coralloid Roots of Cultivated Cycas revoluta (Cycadaceae).Microorganisms. 2023 Sep 21;11(9):2364. doi: 10.3390/microorganisms11092364. Microorganisms. 2023. PMID: 37764208 Free PMC article.
-
Soil Microbial Community Characteristics and Their Effect on Tea Quality under Different Fertilization Treatments in Two Tea Plantations.Genes (Basel). 2024 May 11;15(5):610. doi: 10.3390/genes15050610. Genes (Basel). 2024. PMID: 38790239 Free PMC article.
-
Endosymbiotic Fungal Diversity and Dynamics of the Brown Planthopper across Developmental Stages, Tissues, and Sexes Revealed Using Circular Consensus Sequencing.Insects. 2024 Jan 29;15(2):87. doi: 10.3390/insects15020087. Insects. 2024. PMID: 38392507 Free PMC article.
-
Effects of bacterial fertilizer and soil amendment on Spuriopinella brachycarpa (Kom.) Kitag. growth and soil microbiota.Front Microbiol. 2025 Apr 25;16:1560982. doi: 10.3389/fmicb.2025.1560982. eCollection 2025. Front Microbiol. 2025. PMID: 40351309 Free PMC article.
References
-
- Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196-1206. - PubMed
-
- Bulgarelli D, Rott M, Schlaeppi K, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91-95. - PubMed
-
- Hacquard S, Garrido-Oter R, Gonzales A, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 2015;17:603-616. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

