Electrocardiographic Imaging: A Comparison of Iterative Solvers
- PMID: 33613311
- PMCID: PMC7886787
- DOI: 10.3389/fphys.2021.620250
Electrocardiographic Imaging: A Comparison of Iterative Solvers
Abstract
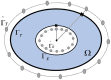
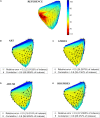
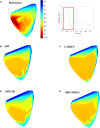
Cardiac disease is a leading cause of morbidity and mortality in developed countries. Currently, non-invasive techniques that can identify patients at risk and provide accurate diagnosis and ablation guidance therapy are under development. One of these is electrocardiographic imaging (ECGI). In ECGI, the first step is to formulate a forward problem that relates the unknown potential sources on the cardiac surface to the measured body surface potentials. Then, the unknown potential sources on the cardiac surface are reconstructed through the solution of an inverse problem. Unfortunately, ECGI still lacks accuracy due to the underlying inverse problem being ill-posed, and this consequently imposes limitations on the understanding and treatment of many cardiac diseases. Therefore, it is necessary to improve the solution of the inverse problem. In this work, we transfer and adapt four inverse problem methods to the ECGI setting: algebraic reconstruction technique (ART), random ART, ART Split Bregman (ART-SB) and range restricted generalized minimal residual (RRGMRES) method. We test all these methods with data from the Experimental Data and Geometric Analysis Repository (EDGAR) and compare their solution with the recorded epicardial potentials provided by EDGAR and a generalized minimal residual (GMRES) iterative method computed solution. Activation maps are also computed and compared. The results show that ART achieved the most stable solutions and, for some datasets, returned the best reconstruction. Differences between the solutions derived from ART and random ART are almost negligible, and the accuracy of their solutions is followed by RRGMRES, ART-SB and finally the GMRES (which returned the worst reconstructions). The RRGMRES method provided the best reconstruction for some datasets but appeared to be less stable than ART when comparing different datasets. In conclusion, we show that the proposed methods (ART, random ART, and RRGMRES) improve the GMRES solution, which has been suggested as inverse problem solution for ECGI.
Keywords: ART; ART-SB; ECGI; GMRES; MFS; RRGMRES; inverse problem; iterative methods.
Copyright © 2021 Borràs and Chamorro-Servent.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

