The Structure of Rhizosphere Fungal Communities of Wild and Domesticated Rice: Changes in Diversity and Co-occurrence Patterns
- PMID: 33613482
- PMCID: PMC7890246
- DOI: 10.3389/fmicb.2021.610823
The Structure of Rhizosphere Fungal Communities of Wild and Domesticated Rice: Changes in Diversity and Co-occurrence Patterns
Abstract
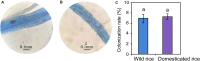
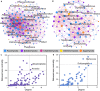
The rhizosphere fungal community affects the ability of crops to acquire nutrients and their susceptibility to pathogen invasion. However, the effects of rice domestication on the diversity and interactions of rhizosphere fungal community still remain largely unknown. Here, internal transcribed spacer amplicon sequencing was used to systematically analyze the structure of rhizosphere fungal communities of wild and domesticated rice. The results showed that domestication increased the alpha diversity indices of the rice rhizosphere fungal community. The changes of alpha diversity index may be associated with the enrichment of Acremonium, Lecythophora, and other specific rare taxa in the rhizosphere of domesticated rice. The co-occurrence network showed that the complexity of wild rice rhizosphere fungal community was higher than that of the domesticated rice rhizosphere fungal community. Arbuscular mycorrhizal fungi (AMF) and soilborne fungi were positively and negatively correlated with more fungi in the wild rice rhizosphere, respectively. For restructuring the rhizomicrobial community of domesticated crops, we hypothesize that microbes that hold positive connections with AMF and negative connections with soilborne fungi can be used as potential sources for bio-inoculation. Our findings provide a scientific basis for reshaping the structure of rhizomicrobial community and furthermore create potential for novel intelligent and sustainable agricultural solutions.
Keywords: arbuscular mycorrhizal fungi; co-occurrence patterns; domestication; rhizosphere; soil-borne fungi.
Copyright © 2021 Chang, Sun, Tian, Ji, Luo, Nasir, Kuramae and Tian.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bin Rahman A. N. M. R., Zhang J. (2016). Flood and drought tolerance in rice: opposite but may coexist. Food Energy Secur. 5 76–88. 10.1002/fes3.79 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

