The Orientia tsutsugamushi ScaB Autotransporter Protein Is Required for Adhesion and Invasion of Mammalian Cells
- PMID: 33613493
- PMCID: PMC7890071
- DOI: 10.3389/fmicb.2021.626298
The Orientia tsutsugamushi ScaB Autotransporter Protein Is Required for Adhesion and Invasion of Mammalian Cells
Abstract
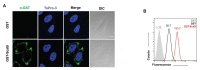
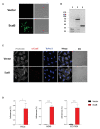
Autotransporter proteins are widely present in Gram-negative bacteria. They play a pivotal role in processes related to bacterial pathogenesis, including adhesion, invasion, colonization, biofilm formation, and cellular toxicity. Bioinformatics analysis revealed that Orientia tsutsugamushi, the causative agent of scrub typhus, encodes six different autotransporter genes (scaA-scaF). Although four of these genes (scaA, scaC, scaD, and scaE) are present in diverse strains, scaB and scaF have been detected in only a limited number of strains. Previous studies have demonstrated that ScaA and ScaC are involved in the adherence of host cells. However, the putative function of other O. tsutsugamushi Sca proteins has not been studied yet. In this study, we show that scaB is transcribed and expressed on the surface of O. tsutsugamushi Boryong strain. Using a heterologous Escherichia coli expression system, we demonstrated that ScaB-expressing E. coli can successfully mediate adherence to and invasion into non-phagocytic cells, including epithelial and endothelial cells. In addition, pretreatment with a recombinant ScaB polypeptide inhibits the entry of O. tsutsugamushi into cultured mammalian cells. Finally, we also identified the scaB gene in the Kuroki and TA686 strains and observed high levels of sequence variation in the passenger domains. Here, we propose that the ScaB protein of O. tsutsugamushi can mediate both adhesion to and invasion into host cells in the absence of other O. tsutsugamushi genes and may play important roles in bacterial pathogenesis.
Keywords: adhesion; autotransporter; invasion; scrub typhus; vaccine.
Copyright © 2021 Nguyen, Kim, Kim, Jeon, Kim, Ha and Cho.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
An Alternative Splicing Variant of the Mixed-Lineage Leukemia 5 Protein Is a Cellular Adhesion Receptor for ScaA of Orientia tsutsugamushi.mBio. 2023 Feb 28;14(1):e0154322. doi: 10.1128/mbio.01543-22. Epub 2022 Dec 21. mBio. 2023. PMID: 36541760 Free PMC article.
-
Detection and distribution of Sca autotransporter protein antigens in diverse isolates of Orientia tsutsugamushi.PLoS Negl Trop Dis. 2018 Sep 20;12(9):e0006784. doi: 10.1371/journal.pntd.0006784. eCollection 2018 Sep. PLoS Negl Trop Dis. 2018. PMID: 30235210 Free PMC article.
-
An Inducible Expression System for Recombinant Sca Proteins with an Autotransporter Domain from Orientia Tsutsugamushi in Escherichia coli.Protein Pept Lett. 2021;28(3):241-248. doi: 10.2174/0929866527666200924144908. Protein Pept Lett. 2021. PMID: 32972336
-
An autotransporter protein from Orientia tsutsugamushi mediates adherence to nonphagocytic host cells.Infect Immun. 2011 Apr;79(4):1718-27. doi: 10.1128/IAI.01239-10. Epub 2011 Jan 31. Infect Immun. 2011. PMID: 21282412 Free PMC article.
-
Scrub Typhus Pathogenesis: Innate Immune Response and Lung Injury During Orientia tsutsugamushi Infection.Front Microbiol. 2019 Sep 6;10:2065. doi: 10.3389/fmicb.2019.02065. eCollection 2019. Front Microbiol. 2019. PMID: 31555249 Free PMC article. Review.
Cited by
-
An atlas of human vector-borne microbe interactions reveals pathogenicity mechanisms.Cell. 2024 Jul 25;187(15):4113-4127.e13. doi: 10.1016/j.cell.2024.05.023. Epub 2024 Jun 13. Cell. 2024. PMID: 38876107 Free PMC article.
-
A scrub typhus vaccine presents a challenging unmet need.NPJ Vaccines. 2023 Feb 9;8(1):11. doi: 10.1038/s41541-023-00605-1. NPJ Vaccines. 2023. PMID: 36759505 Free PMC article. Review.
-
Moonlighting in Rickettsiales: Expanding Virulence Landscape.Trop Med Infect Dis. 2022 Feb 19;7(2):32. doi: 10.3390/tropicalmed7020032. Trop Med Infect Dis. 2022. PMID: 35202227 Free PMC article. Review.
-
Analysis of Orientia tsutsugamushi promoter activity.Pathog Dis. 2021 Sep 23;79(7):ftab044. doi: 10.1093/femspd/ftab044. Pathog Dis. 2021. PMID: 34515306 Free PMC article.
-
An Alternative Splicing Variant of the Mixed-Lineage Leukemia 5 Protein Is a Cellular Adhesion Receptor for ScaA of Orientia tsutsugamushi.mBio. 2023 Feb 28;14(1):e0154322. doi: 10.1128/mbio.01543-22. Epub 2022 Dec 21. mBio. 2023. PMID: 36541760 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

