Co-Occurrence of Listeria spp. and Spoilage Associated Microbiota During Meat Processing Due to Cross-Contamination Events
- PMID: 33613505
- PMCID: PMC7892895
- DOI: 10.3389/fmicb.2021.632935
Co-Occurrence of Listeria spp. and Spoilage Associated Microbiota During Meat Processing Due to Cross-Contamination Events
Abstract
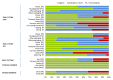
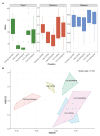
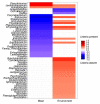
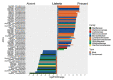
A large part of foodborne outbreaks related to Listeria monocytogenes are linked to meat and meat products. Especially, recontamination of meat products and deli-meat during slicing, packaging, and repackaging is in the focus of food authorities. In that regard, L. monocytogenes persistence in multi-species biofilms is one major issue, since they survive elaborate cleaning and disinfection measures. Here, we analyzed the microbial community structure throughout a meat processing facility using a combination of high-throughput full-length 16S ribosomal RNA (rRNA) gene sequencing and traditional microbiological methods. Samples were taken at different stages during meat cutting as well as from multiple sites throughout the facility environment to capture the product and the environmental associated microbiota co-occurring with Listeria spp. and L. monocytogenes. The listeria testing revealed a widely disseminated contamination (50%; 88 of 176 samples were positive for Listeria spp. and 13.6%; 24 of 176 samples were positive for L. monocytogenes). The pulsed-field gel electrophoresis (PFGE) typing evidenced 14 heterogeneous L. monocytogenes profiles with PCR-serogroup 1/2a, 3a as most dominant. PFGE type MA3-17 contributed to the resilient microbiota of the facility environment and was related to environmental persistence. The core in-house microbiota consisted mainly of the genera Acinetobacter, Pseudomonas, Psychrobacter (Proteobacteria), Anaerobacillus, Bacillus (Firmicutes), and Chryseobacterium (Bacteroidota). While the overall microbial community structure clearly differed between product and environmental samples, we were able to discern correlation patterns regarding the presence/absence of Listeria spp. in both sample groups. Specifically, our longitudinal analysis revealed association of Listeria spp. with known biofilm-producing Pseudomonas, Acinetobacter, and Janthinobacterium species on the meat samples. Similar patterns were also observed on the surface, indicating dispersal of microorganisms from this multispecies biofilm. Our data provided a better understanding of the built environment microbiome in the meat processing context and promoted more effective options for targeted disinfection in the analyzed facility.
Keywords: Listeria monocytogenes; meat processing; microbial communities; microbiome; spoilage.
Copyright © 2021 Zwirzitz, Wetzels, Dixon, Fleischmann, Selberherr, Thalguter, Quijada, Dzieciol, Wagner and Stessl.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Allam M., Tau N., Smouse S. L., Mtshali P. S., Mnyameni F., Khumalo Z. T. H., et al. (2018). Whole-genome sequences of Listeria monocytogenes sequence type 6 isolates associated with a large foodborne outbreak in South Africa, 2017 to 2018. Genome Announc. 6:e00538–18. 10.1128/genomeA.00538-18, PMID: - DOI - PMC - PubMed
-
- Andersen K. S., Kirkegaard R. H., Karst S. M., Albertsen M. (2018). “Ampvis2 An R Package to Analyse and Visualise 16S RRNA Amplicon Data.” BioRxiv [Preprint]. 299537. 10.1101/299537 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

