Characterization of Mungbean CONSTANS-LIKE Genes and Functional Analysis of CONSTANS-LIKE 2 in the Regulation of Flowering Time in Arabidopsis
- PMID: 33613600
- PMCID: PMC7890258
- DOI: 10.3389/fpls.2021.608603
Characterization of Mungbean CONSTANS-LIKE Genes and Functional Analysis of CONSTANS-LIKE 2 in the Regulation of Flowering Time in Arabidopsis
Abstract
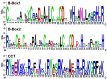
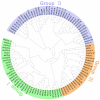
CONSTANS-LIKE (COL) genes play important roles in the regulation of plant growth and development, and they have been analyzed in many plant species. However, few studies have examined COL genes in mungbean (Vigna radiata). In this study, we identified and characterized 31 mungbean genes whose proteins contained B-Box domains. Fourteen were designated as VrCOL genes and were distributed on 7 of the 11 mungbean chromosomes. Based on their phylogenetic relationships, VrCOLs were clustered into three groups (I, II, and III), which contained 4, 6, and 4 members, respectively. The gene structures and conserved motifs of the VrCOL genes were analyzed, and two duplicated gene pairs, VrCOL1/VrCOL2 and VrCOL8/VrCOL9, were identified. A total of 82 cis-acting elements were found in the VrCOL promoter regions, and the numbers and types of cis-acting elements in each VrCOL promoter region differed. As a result, the expression patterns of VrCOLs varied in different tissues and throughout the day under long-day and short-day conditions. Among these VrCOL genes, VrCOL2 showed a close phylogenetic relationship with Arabidopsis thaliana CO and displayed daily oscillations in expression under short-day conditions but not long-day conditions. In addition, overexpression of VrCOL2 accelerated flowering in Arabidopsis under short-day conditions by affecting the expression of the flowering time genes AtFT and AtTSF. Our study lays the foundation for further investigation of VrCOL gene functions.
Keywords: CONSTANS; VrCOL2; flowering time; genome wide; mungbean.
Copyright © 2021 Liu, Zhang, Zhu, Cai and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Genome-wide identification and characterization of mungbean CIRCADIAN CLOCK ASSOCIATED 1 like genes reveals an important role of VrCCA1L26 in flowering time regulation.BMC Genomics. 2022 May 17;23(1):374. doi: 10.1186/s12864-022-08620-7. BMC Genomics. 2022. PMID: 35581536 Free PMC article.
-
Mungbean DIRIGENT Gene Subfamilies and Their Expression Profiles Under Salt and Drought Stresses.Front Genet. 2021 Sep 22;12:658148. doi: 10.3389/fgene.2021.658148. eCollection 2021. Front Genet. 2021. PMID: 34630501 Free PMC article.
-
Expression Analysis and Regulation Network Identification of the CONSTANS-Like Gene Family in Moso Bamboo (Phyllostachys edulis) Under Photoperiod Treatments.DNA Cell Biol. 2019 Jul;38(7):607-626. doi: 10.1089/dna.2018.4611. Epub 2019 Jun 17. DNA Cell Biol. 2019. PMID: 31210530
-
Genome-wide identification and characterization of CONSTANS-like gene family in radish (Raphanus sativus).PLoS One. 2018 Sep 24;13(9):e0204137. doi: 10.1371/journal.pone.0204137. eCollection 2018. PLoS One. 2018. PMID: 30248137 Free PMC article.
-
The regulation of flowering time by daylength in Arabidopsis.Symp Soc Exp Biol. 1998;51:105-10. Symp Soc Exp Biol. 1998. PMID: 10645431 Review.
Cited by
-
Transcriptome Analysis of Flower Development and Mining of Genes Related to Flowering Time in Tomato (Solanum lycopersicum).Int J Mol Sci. 2021 Jul 29;22(15):8128. doi: 10.3390/ijms22158128. Int J Mol Sci. 2021. PMID: 34360893 Free PMC article.
-
Involvement of CONSTANS-like Proteins in Plant Flowering and Abiotic Stress Response.Int J Mol Sci. 2023 Nov 22;24(23):16585. doi: 10.3390/ijms242316585. Int J Mol Sci. 2023. PMID: 38068908 Free PMC article. Review.
-
Genome-wide exploration of the CONSTANS-like (COL) gene family and its potential role in regulating plant flowering time in foxtail millet (Setaria italica).Sci Rep. 2024 Oct 18;14(1):24518. doi: 10.1038/s41598-024-74724-7. Sci Rep. 2024. PMID: 39424865 Free PMC article.
-
Genome-wide identification and characterization of mungbean CIRCADIAN CLOCK ASSOCIATED 1 like genes reveals an important role of VrCCA1L26 in flowering time regulation.BMC Genomics. 2022 May 17;23(1):374. doi: 10.1186/s12864-022-08620-7. BMC Genomics. 2022. PMID: 35581536 Free PMC article.
-
Characterization of Phytohormones and Transcriptomic Profiling of the Female and Male Inflorescence Development in Manchurian Walnut (Juglans mandshurica Maxim.).Int J Mol Sci. 2022 May 13;23(10):5433. doi: 10.3390/ijms23105433. Int J Mol Sci. 2022. PMID: 35628244 Free PMC article.
References
-
- Beinecke F. A., Grundmann L., Wiedmann D. R., Schmidt F. J., Caesar A. S., Zimmermann M., et al. (2018). The FT/FD-dependent initiation of flowering under long-day conditions in the day-neutral species Nicotiana tabacum originates from the facultative short-day ancestor Nicotiana tomentosiformis. Plant J. 96 329–342. 10.1111/tpj.14033 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

