The Genome of Banana Leaf Blight Pathogen Fusarium sacchari str. FS66 Harbors Widespread Gene Transfer From Fusarium oxysporum
- PMID: 33613610
- PMCID: PMC7889605
- DOI: 10.3389/fpls.2021.629859
The Genome of Banana Leaf Blight Pathogen Fusarium sacchari str. FS66 Harbors Widespread Gene Transfer From Fusarium oxysporum
Abstract
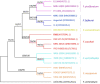
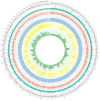
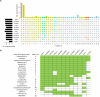
Fusarium species have been identified as pathogens causing many different plant diseases, and here we report an emerging banana leaf blight (BLB) caused by F. sacchari (Fs) discovered in Guangdong, China. From the symptomatic tissues collected in the field, a fungal isolate was obtained, which induced similar symptoms on healthy banana seedlings after inoculation. Koch's postulates were fulfilled after the re-isolation of the pathogen. Phylogenetic analysis on two gene segments and the whole genome sequence identified the pathogen belonging to Fs and named as Fs str. FS66. A 45.74 Mb genome of FS66 was acquired through de novo assembly using long-read sequencing data, and its contig N50 (1.97 Mb) is more than 10-fold larger than the previously available genome in the species. Based on transcriptome sequencing and ab initio gene annotation, a total of 14,486 protein-encoding genes and 418 non-coding RNAs were predicted. A total of 48 metabolite biosynthetic gene clusters including the fusaric acid biosynthesis gene cluster were predicted in silico in the FS66 genome. Comparison between FS66 and other 11 Fusarium genomes identified tens to hundreds of genes specifically gained and lost in FS66, including some previously correlated with Fusarium pathogenicity. The FS66 genome also harbors widespread gene transfer on the core chromosomes putatively from F. oxysporum species complex (FOSC), including 30 involved in Fusarium pathogenicity/virulence. This study not only reports the BLB caused by Fs, but also provides important information and clues for further understanding of the genome evolution among pathogenic Fusarium species.
Keywords: Fusarium sacchari; banana leaf blight; comparative genomics; de novo assembly; gene transfer.
Copyright © 2021 Cui, Wu, Peng, Song and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Araújo N. A. F., Pasqual M., Pio L. A. S., Alves E., de Matos Moura N., Costa S. D. S. (2017). Identification and aggressiveness of four isolates of Fusarium oxysporumf. sp. cubense from Latundan banana in Brazil. J. Phytopathol. 165 257–264. 10.1111/jph.12557 - DOI
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

