Resolving Recalcitrant Clades in the Pantropical Ochnaceae: Insights From Comparative Phylogenomics of Plastome and Nuclear Genomic Data Derived From Targeted Sequencing
- PMID: 33613613
- PMCID: PMC7890083
- DOI: 10.3389/fpls.2021.638650
Resolving Recalcitrant Clades in the Pantropical Ochnaceae: Insights From Comparative Phylogenomics of Plastome and Nuclear Genomic Data Derived From Targeted Sequencing
Abstract
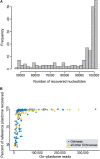
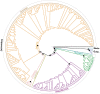
Plastid DNA sequence data have been traditionally widely used in plant phylogenetics because of the high copy number of plastids, their uniparental inheritance, and the blend of coding and non-coding regions with divergent substitution rates that allow the reconstruction of phylogenetic relationships at different taxonomic ranks. In the present study, we evaluate the utility of the plastome for the reconstruction of phylogenetic relationships in the pantropical plant family Ochnaceae (Malpighiales). We used the off-target sequence read fraction of a targeted sequencing study (targeting nuclear loci only) to recover more than 100 kb of the plastid genome from the majority of the more than 200 species of Ochnaceae and all but two genera using de novo and reference-based assembly strategies. Most of the recalcitrant nodes in the family's backbone were resolved by our plastome-based phylogenetic inference, corroborating the most recent classification system of Ochnaceae and findings from a phylogenomic study based on nuclear loci. Nonetheless, the phylogenetic relationships within the major clades of tribe Ochnineae, which comprise about two thirds of the family's species diversity, received mostly low support. Generally, the phylogenetic resolution was lowest at the infrageneric level. Overall there was little phylogenetic conflict compared to a recent analysis of nuclear loci. Effects of taxon sampling were invoked as the most likely reason for some of the few well-supported discords. Our study demonstrates the utility of the off-target fraction of a target enrichment study for assembling near-complete plastid genomes for a large proportion of samples.
Keywords: Malpighiales; hybrid enrichment; off-target reads; phylogenetic conflict; phylogenomics; plastome; taxon sampling.
Copyright © 2021 Schneider, Paule, Jungcurt, Cardoso, Amorim, Berberich and Zizka.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Amaral M. C. E. (1991). Phylogenetische Systematik der Ochnaceae. Bot. Jahrb. Syst. 113 105–196.
-
- Amaral M. C. E., Bittrich V. (2014). “Ochnaceae,” in The Families and Genera of Vascular Plants, Vol. 11 ed. Kubitzki K. (Heidelberg: Springer; ), 253–268. 10.1007/978-3-642-39417-1_19 - DOI
-
- Bakker F. T. (2017). Herbarium genomics: skimming and plastomics from archival specimens. Webbia 72 35–45. 10.1080/00837792.2017.1313383 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

