Identification of a Metastasis-Associated Gene Signature of Clear Cell Renal Cell Carcinoma
- PMID: 33613617
- PMCID: PMC7889952
- DOI: 10.3389/fgene.2020.603455
Identification of a Metastasis-Associated Gene Signature of Clear Cell Renal Cell Carcinoma
Abstract
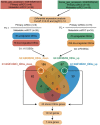
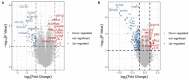
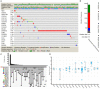
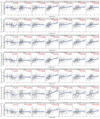
Clear cell renal cell carcinoma (ccRCC) is one of the most frequent pathological subtypes of kidney cancer, accounting for ~70-75%, and the major cause of mortality is metastatic disease. The difference in gene expression profiles between primary ccRCC tumors and metastatic tumors has not been determined. Thus, we report integrated genomic and transcriptomic analysis for identifying differentially expressed genes (DEGs) between primary and metastatic ccRCC tumors to understand the molecular mechanisms underlying the development of metastases. The microarray datasets GSE105261 and GSE85258 were obtained from the Gene Expression Omnibus (GEO) database, and the R package limma was used for DEG analyses. In summary, the results described herein provide important molecular evidence that metastatic ccRCC tumors are different from primary tumors. Enrichment analysis indicated that the DEGs were mainly enriched in ECM-receptor interaction, platelet activation, protein digestion, absorption, focal adhesion, and the PI3K-Akt signaling pathway. Moreover, we found that DEGs associated with a higher level of tumor immune infiltrates and tumor mutation burden were more susceptible to poor prognosis of ccRCC. Specifically, our study indicates that seven core genes, namely the collagen family (COL1A2, COL1A1, COL6A3, and COL5A1), DCN, FBLN1, and POSTN, were significantly upregulated in metastatic tumors compared with those in primary tumors and, thus, potentially offer insight into novel therapeutic and early diagnostic biomarkers of ccRCC.
Keywords: biomarker; clear cell renal cell carcinoma; immune infiltration; metastasis; tumor mutation burden.
Copyright © 2021 Gao, Yan, Zhang, Fan, Jiao and Shao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

