Genetic Diversity and Signatures of Selection for Thermal Stress in Cattle and Other Two Bos Species Adapted to Divergent Climatic Conditions
- PMID: 33613634
- PMCID: PMC7887320
- DOI: 10.3389/fgene.2021.604823
Genetic Diversity and Signatures of Selection for Thermal Stress in Cattle and Other Two Bos Species Adapted to Divergent Climatic Conditions
Abstract
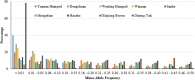
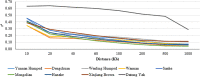
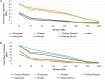
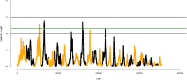
Understanding the biological mechanisms of climatic adaptation is of paramount importance for the optimization of breeding programs and conservation of genetic resources. The aim of this study was to investigate genetic diversity and unravel genomic regions potentially under selection for heat and/or cold tolerance in thirty-two worldwide cattle breeds, with a focus on Chinese local cattle breeds adapted to divergent climatic conditions, Datong yak (Bos grunniens; YAK), and Bali (Bos javanicus) based on dense SNP data. In general, moderate genetic diversity levels were observed in most cattle populations. The proportion of polymorphic SNP ranged from 0.197 (YAK) to 0.992 (Mongolian cattle). Observed and expected heterozygosity ranged from 0.023 (YAK) to 0.366 (Sanhe cattle; SH), and from 0.021 (YAK) to 0.358 (SH), respectively. The overall average inbreeding (±SD) was: 0.118 ± 0.028, 0.228 ± 0.059, 0.194 ± 0.041, and 0.021 ± 0.004 based on the observed versus expected number of homozygous genotypes, excess of homozygosity, correlation between uniting gametes, and runs of homozygosity (ROH), respectively. Signatures of selection based on multiple scenarios and methods (F ST, HapFLK, and ROH) revealed important genomic regions and candidate genes. The candidate genes identified are related to various biological processes and pathways such as heat-shock proteins, oxygen transport, anatomical traits, mitochondrial DNA maintenance, metabolic activity, feed intake, carcass conformation, fertility, and reproduction. This highlights the large number of biological processes involved in thermal tolerance and thus, the polygenic nature of climatic resilience. A comprehensive description of genetic diversity measures in Chinese cattle and YAK was carried out and compared to 24 worldwide cattle breeds to avoid potential biases. Numerous genomic regions under positive selection were detected using three signature of selection methods and candidate genes potentially under positive selection were identified. Enriched function analyses pinpointed important biological pathways, molecular function and cellular components, which contribute to a better understanding of the biological mechanisms underlying thermal tolerance in cattle. Based on the large number of genomic regions identified, thermal tolerance has a complex polygenic inheritance nature, which was expected considering the various mechanisms involved in thermal stress response.
Keywords: climate resilience; cold stress; genetic resources; heat stress; heat tolerance; selective sweep.
Copyright © 2021 Freitas, Wang, Yan, Oliveira, Schenkel, Zhang, Xu and Brito.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Whole-genome resequencing reveals genetic diversity, differentiation, and selection signatures of yak breeds/populations in Qinghai, China.Front Genet. 2023 Jan 10;13:1034094. doi: 10.3389/fgene.2022.1034094. eCollection 2022. Front Genet. 2023. PMID: 36704337 Free PMC article.
-
Characterization of runs of homozygosity, heterozygosity-enriched regions, and population structure in cattle populations selected for different breeding goals.BMC Genomics. 2022 Mar 16;23(1):209. doi: 10.1186/s12864-022-08384-0. BMC Genomics. 2022. PMID: 35291953 Free PMC article.
-
Evaluating genomic inbreeding of two Chinese yak (Bos grunniens) populations.BMC Genomics. 2024 Jul 24;25(1):712. doi: 10.1186/s12864-024-10640-4. BMC Genomics. 2024. PMID: 39044139 Free PMC article.
-
Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits.BMC Genomics. 2021 Oct 15;22(1):747. doi: 10.1186/s12864-021-08042-x. BMC Genomics. 2021. PMID: 34654366 Free PMC article. Review.
-
Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data.BMC Genomics. 2021 Jan 9;22(1):43. doi: 10.1186/s12864-020-07340-0. BMC Genomics. 2021. PMID: 33421990 Free PMC article. Review.
Cited by
-
Insights into trait-association of selection signatures and adaptive eQTL in indigenous African cattle.BMC Genomics. 2024 Oct 19;25(1):981. doi: 10.1186/s12864-024-10852-8. BMC Genomics. 2024. PMID: 39425030 Free PMC article.
-
Candidate genes associated with heat stress and breeding strategies to relieve its effects in dairy cattle: a deeper insight into the genetic architecture and immune response to heat stress.Front Vet Sci. 2023 Sep 13;10:1151241. doi: 10.3389/fvets.2023.1151241. eCollection 2023. Front Vet Sci. 2023. PMID: 37771947 Free PMC article. Review.
-
Longitudinal genomic analyses of automatically-recorded vaginal temperature in lactating sows under heat stress conditions based on random regression models.Genet Sel Evol. 2023 Dec 21;55(1):95. doi: 10.1186/s12711-023-00868-1. Genet Sel Evol. 2023. PMID: 38129768 Free PMC article.
-
Copy Number Variation Regions Differing in Segregation Patterns Span Different Sets of Genes.Animals (Basel). 2023 Jul 19;13(14):2351. doi: 10.3390/ani13142351. Animals (Basel). 2023. PMID: 37508128 Free PMC article.
-
Heat stress effects on milk yield traits and metabolites and mitigation strategies for dairy cattle breeds reared in tropical and sub-tropical countries.Front Vet Sci. 2023 Jul 7;10:1121499. doi: 10.3389/fvets.2023.1121499. eCollection 2023. Front Vet Sci. 2023. PMID: 37483284 Free PMC article. Review.
References
-
- Abdurehman A. (2019). Physiological and Anatomical Adaptation Characteristics of Borana Cattle to Pastoralist Lowland Environments. Asian J. Biol. Sci. 12 364–372. 10.3923/ajbs.2019.364.372 - DOI
-
- Ajmone-Marsan P., Garcia J. F., Lenstra J. A. (2010). On the origin of cattle: How aurochs became cattle and colonized the world. Evol. Anthropol. 19 148–157. 10.1002/evan.20267 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

