Stratification of Estrogen Receptor-Negative Breast Cancer Patients by Integrating the Somatic Mutations and Transcriptomic Data
- PMID: 33613637
- PMCID: PMC7886807
- DOI: 10.3389/fgene.2021.610087
Stratification of Estrogen Receptor-Negative Breast Cancer Patients by Integrating the Somatic Mutations and Transcriptomic Data
Abstract
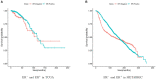
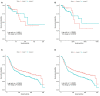
Patients with estrogen receptor-negative breast cancer generally have a worse prognosis than estrogen receptor-positive patients. Nevertheless, a significant proportion of the estrogen receptor-negative cases have favorable outcomes. Identifying patients with a good prognosis, however, remains difficult, as recent studies are quite limited. The identification of molecular biomarkers is needed to better stratify patients. The significantly mutated genes may be potentially used as biomarkers to identify the subtype and to predict outcomes. To identify the biomarkers of receptor-negative breast cancer among the significantly mutated genes, we developed a workflow to screen significantly mutated genes associated with the estrogen receptor in breast cancer by a gene coexpression module. The similarity matrix was calculated with distance correlation to obtain gene modules through a weighted gene coexpression network analysis. The modules highly associated with the estrogen receptor, called important modules, were enriched for breast cancer-related pathways or disease. To screen significantly mutated genes, a new gene list was obtained through the overlap of the important module genes and the significantly mutated genes. The genes on this list can be used as biomarkers to predict survival of estrogen receptor-negative breast cancer patients. Furthermore, we selected six hub significantly mutated genes in the gene list which were also able to separate these patients. Our method provides a new and alternative method for integrating somatic gene mutations and expression data for patient stratification of estrogen receptor-negative breast cancers.
Keywords: breast cancer patient stratification; distance correlation; estrogen receptor-negative; gene coexpression network; significantly mutated gene.
Copyright © 2021 Hou, Ye, Wang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. 10.1158/2159-8290.CD-12-0095 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

