An E2F1/DDX11/EZH2 Positive Feedback Loop Promotes Cell Proliferation in Hepatocellular Carcinoma
- PMID: 33614480
- PMCID: PMC7892623
- DOI: 10.3389/fonc.2020.593293
An E2F1/DDX11/EZH2 Positive Feedback Loop Promotes Cell Proliferation in Hepatocellular Carcinoma
Abstract
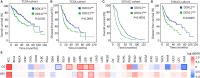
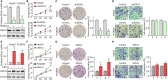
Hepatocellular carcinoma (HCC) accounts for one of the leading causes of cancer-related death, and is attributed to the dysregulation of genes involved in genome stability. DDX11, a DNA helicase, has been implicated in rare genetic disease and human cancers. Yet, its clinical value, biological function, and the underlying mechanism in HCC progression are not fully understood. Here, we show that DDX11 is upregulated in HCC and exhibits oncogenic activity via EZH2/p21 signaling. High expression of DDX11 is significantly correlated with poor outcomes of HCC patients in two independent cohorts. DDX11 overexpression increases HCC cell viabilities and colony formation, whereas DDX11 knockdown arrests cells at G1 phase without alteration of p53 expression. Ectopic expression of DDX11 reduces, while depletion of DDX11 induces the expression of p21. Treatment of p21 siRNA markedly attenuates the cell growth suppression caused by DDX11 silence. Further studies reveal that DDX11 interacts with EZH2 in HCC cells to protect it from ubiquitination-mediated protein degradation, consequently resulting in the downregulation of p21. In addition, E2F1 is identified as one of the upstream regulators of DDX11, and forms a positive feedback loop with EZH2 to upregulate DDX11 and facilitate cell proliferation. Collectively, our data suggest DDX11 as a promising prognostic factor and an oncogene in HCC via a E2F1/DDX11/EZH2 positive feedback loop.
Keywords: DDX11; E2F1; EZH2; hepatocellular carcinoma; p21.
Copyright © 2021 Su, Li, Zhang, Zhang, Shen and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Long noncoding RNA DDX11-AS1 epigenetically represses LATS2 by interacting with EZH2 and DNMT1 in hepatocellular carcinoma.Biochem Biophys Res Commun. 2019 Jul 5;514(4):1051-1057. doi: 10.1016/j.bbrc.2019.05.042. Epub 2019 May 13. Biochem Biophys Res Commun. 2019. PMID: 31097223
-
LINC00978 promotes the progression of hepatocellular carcinoma by regulating EZH2-mediated silencing of p21 and E-cadherin expression.Cell Death Dis. 2019 Oct 3;10(10):752. doi: 10.1038/s41419-019-1990-6. Cell Death Dis. 2019. PMID: 31582742 Free PMC article.
-
FBXO22 promotes the development of hepatocellular carcinoma by regulating the ubiquitination and degradation of p21.J Exp Clin Cancer Res. 2019 Feb 26;38(1):101. doi: 10.1186/s13046-019-1058-6. J Exp Clin Cancer Res. 2019. PMID: 30808376 Free PMC article.
-
UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53.Biochem Biophys Res Commun. 2017 Nov 4;493(1):20-27. doi: 10.1016/j.bbrc.2017.09.091. Epub 2017 Sep 18. Biochem Biophys Res Commun. 2017. PMID: 28935368
-
The Genome Stability Maintenance DNA Helicase DDX11 and Its Role in Cancer.Genes (Basel). 2021 Mar 10;12(3):395. doi: 10.3390/genes12030395. Genes (Basel). 2021. PMID: 33802088 Free PMC article. Review.
Cited by
-
Construction of a radiogenomic signature based on endoplasmic reticulum stress for predicting prognosis and systemic combination therapy response in hepatocellular carcinoma.BMC Cancer. 2025 Jan 23;25(1):131. doi: 10.1186/s12885-025-13433-4. BMC Cancer. 2025. PMID: 39849389 Free PMC article.
-
Identification of the E2F1-RAD51AP1 axis as a key factor in MGMT-methylated GBM TMZ resistance.Cancer Biol Med. 2023 Jun 5;20(5):385-400. doi: 10.20892/j.issn.2095-3941.2023.0011. Cancer Biol Med. 2023. PMID: 37283490 Free PMC article.
-
Piperlongumine inhibits renal cell carcinoma progression by modulating the DDX11-miR-15b-3p-DLD axis.Transl Androl Urol. 2025 Apr 30;14(4):897-912. doi: 10.21037/tau-2025-11. Epub 2025 Apr 27. Transl Androl Urol. 2025. PMID: 40376520 Free PMC article.
-
E2F1 combined with LINC01004 super-enhancer to promote hepatocellular carcinoma cell proliferation and metastasis.Clin Epigenetics. 2023 Jan 31;15(1):17. doi: 10.1186/s13148-023-01428-6. Clin Epigenetics. 2023. PMID: 36721155 Free PMC article.
-
Identification of Prognosis-Related Oxidative Stress Model with Immunosuppression in HCC.Biomedicines. 2023 Feb 24;11(3):695. doi: 10.3390/biomedicines11030695. Biomedicines. 2023. PMID: 36979675 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

