Identification and Validation of an Autophagy-Related lncRNA Signature for Patients With Breast Cancer
- PMID: 33614483
- PMCID: PMC7892971
- DOI: 10.3389/fonc.2020.597569
Identification and Validation of an Autophagy-Related lncRNA Signature for Patients With Breast Cancer
Abstract
Background: Autophagy is a "self-feeding" phenomenon of cells, which is crucial in mammalian development. Long non-coding RNA (lncRNA) is a new regulatory factor for cell autophagy, which can regulate the process of autophagy to affect tumor progression. However, poor attention has been paid to the roles of autophagy-related lncRNAs in breast cancer.
Objective: This study aimed to construct an autophagy-related lncRNA signature that can effectively predict the prognosis of breast cancer patients and explore the potential functions of these lncRNAs.
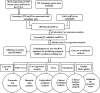
Methods: The RNA sequencing (RNA-Seq) data of breast cancer patients was collected from The Cancer Genome Atlas (TCGA) database and the GSE20685 database. Multivariate Cox analysis was implemented to produce an autophagy-related lncRNA signature in the TCGA cohort. The signature was then validated in the GSE20685 cohort. The receiver operator characteristic (ROC) curve was performed to evaluate the predictive ability of the signature. Gene set enrichment analysis (GSEA) was used to explore the potential functions based on the signature. Finally, the study developed a nomogram and internal verification based on the autophagy-related lncRNAs.
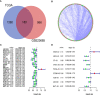
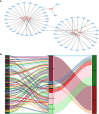
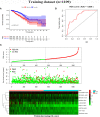
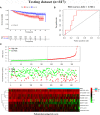
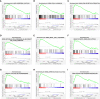
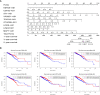
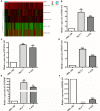
Results: A signature composed of 9 autophagy-related lncRNAs was determined as a prognostic model, and 1,109 breast cancer patients were divided into high-risk group and low-risk group based on median risk score of the signature. Further analysis demonstrated that the over survival (OS) of breast cancer patients in the high-risk group was poorer than that in the low-risk group based on the prognostic signature. The area under the curve (AUC) of ROC curve verified the sensitivity and specificity of this signature. Additionally, we confirmed the signature is an independent factor and found it may be correlated to the progression of breast cancer. GSEA showed gene sets were notably enriched in carcinogenic activation pathways and autophagy-related pathways. The qRT-PCR identified 5 lncRNAs with significantly differential expression in breast cancer cells based on the 9 lncRNAs of the prognostic model, and the results were consistent with the tissues.
Conclusion: In summary, our signature has potential predictive value in the prognosis of breast cancer and these autophagy-related lncRNAs may play significant roles in the diagnosis and treatment of breast cancer.
Keywords: The Cancer Genome Atlas; autophagy; breast cancer; long non-coding RNA; prognostic signature.
Copyright © 2021 Zhang, Zhu, Yin, Yang, Guo, Zhang, Zhou and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Identification and Validation of Six Autophagy-related Long Non-coding RNAs as Prognostic Signature in Colorectal Cancer.Int J Med Sci. 2021 Jan 1;18(1):88-98. doi: 10.7150/ijms.49449. eCollection 2021. Int J Med Sci. 2021. PMID: 33390777 Free PMC article.
-
A Signature of Autophagy-Related Long Non-coding RNA to Predict the Prognosis of Breast Cancer.Front Genet. 2021 Mar 16;12:569318. doi: 10.3389/fgene.2021.569318. eCollection 2021. Front Genet. 2021. PMID: 33796128 Free PMC article.
-
Identification and validation of an autophagy-related long non-coding RNA signature as a prognostic biomarker for patients with lung adenocarcinoma.J Thorac Dis. 2021 Feb;13(2):720-734. doi: 10.21037/jtd-20-2803. J Thorac Dis. 2021. PMID: 33717544 Free PMC article.
-
Critical roles of lncRNA-mediated autophagy in urologic malignancies.Front Pharmacol. 2024 Jun 13;15:1405199. doi: 10.3389/fphar.2024.1405199. eCollection 2024. Front Pharmacol. 2024. PMID: 38939836 Free PMC article. Review.
-
The role of LncRNA-mediated autophagy in cancer progression.Front Cell Dev Biol. 2024 Jun 12;12:1348894. doi: 10.3389/fcell.2024.1348894. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38933333 Free PMC article. Review.
Cited by
-
Over Fifty Years of Life, Death, and Cannibalism: A Historical Recollection of Apoptosis and Autophagy.Int J Mol Sci. 2021 Nov 18;22(22):12466. doi: 10.3390/ijms222212466. Int J Mol Sci. 2021. PMID: 34830349 Free PMC article. Review.
-
High miR-3609 expression is associated with better prognosis in TNBC based on mining using systematic integrated public sequencing data.Exp Ther Med. 2022 Jan;23(1):54. doi: 10.3892/etm.2021.10976. Epub 2021 Nov 16. Exp Ther Med. 2022. PMID: 34934431 Free PMC article.
-
Multi-Omics Marker Analysis Enables Early Prediction of Breast Tumor Progression.Front Genet. 2021 Jun 3;12:670749. doi: 10.3389/fgene.2021.670749. eCollection 2021. Front Genet. 2021. PMID: 34149812 Free PMC article.
-
Identification of an autophagy- and macropinocytosis-related prognostic signature for the prediction of prognosis and therapeutic response in gastric cancer.Genes Genomics. 2024 Oct;46(10):1149-1164. doi: 10.1007/s13258-024-01557-z. Epub 2024 Aug 16. Genes Genomics. 2024. PMID: 39150612
-
Emerging role of lncRNAs in drug resistance mechanisms in head and neck squamous cell carcinoma.Front Oncol. 2022 Aug 1;12:965628. doi: 10.3389/fonc.2022.965628. eCollection 2022. Front Oncol. 2022. PMID: 35978835 Free PMC article. Review.
References
-
- El Sayed R, El Jamal L, El Iskandarani S, Kort J, Abdel Salam M, Assi H. Endocrine and Targeted Therapy for Hormone-Receptor-Positive, HER2-Negative Advanced Breast Cancer: Insights to Sequencing Treatment and Overcoming Resistance Based on Clinical Trials. Front Oncol (2019) 9:510. 10.3389/fonc.2019.00510 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

