Epidemic Trend and Molecular Evolution of HV Family in the Main Hantavirus Epidemic Areas From 2004 to 2016, in P.R. China
- PMID: 33614521
- PMCID: PMC7886990
- DOI: 10.3389/fcimb.2020.584814
Epidemic Trend and Molecular Evolution of HV Family in the Main Hantavirus Epidemic Areas From 2004 to 2016, in P.R. China
Abstract
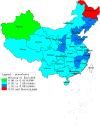
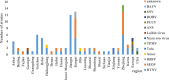
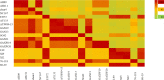
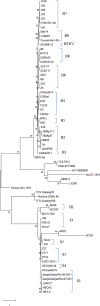
Hemorrhagic fever with renal syndrome (HFRS) is caused by hantavirus (HV) infection, and is prevalent across Europe and Asia (mainly China). The genetic variation and wide host range of the HV family may lead to vaccine failure. In this study, we analyzed the gene sequences of HV isolated from different regions of China in order to trace the molecular evolution of HV and the epidemiological trends of HFRS. A total of 16,6975 HFRS cases and 1,689 HFRS-related deaths were reported from 2004 to 2016, with the average annual incidence rate of 0.9674 per 100,000, 0.0098 per 100,000 mortality rate, and case fatality rate 0.99%. The highest number of cases were detected in 2004 (25,041), and after decreasing to the lowest numbers (8,745) in 2009, showed an incline from 2010. The incidence of HFRS is the highest in spring and winter, and three times as many men are affected as women. In addition, farmers account for the largest proportion of all cases. The main hosts of HV are Rattus norvegicus and Apodemus agrarius, and the SEOV strain is mainly found in R. norvegicus and Niviventer confucianus. Phylogenetic analysis showed that at least 10 HTNV subtypes and 6 SEOV subtypes are endemic to China. We found that the clustering pattern of M genome segments was different from that of the S segments, indicating the possibility of gene recombination across HV strains. The recent increase in the incidence of HFRS may be related to climatic factors, such as temperature, relative humidity and hours of sunshine, as well as biological factors like rodent density, virus load in rodents and genetic variation. The scope of vaccine application should be continuously expanded, and surveillance measures and prevention and control strategies should be improved to reduce HFRS infection in China.
Keywords: epidemical tendency; genetic variation; hantavirus; incidence rate; molecular evolution.
Copyright © 2021 Wang, Yue, Yao, Zhu, Ai, Hu, Zhang, Yang, Yang, Luo, Wang, Hou and Tan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Bai X. H., Peng C., Jiang T., Hu Z. M., Huang D. S., Guan P. (2019). Distribution of geographical scale, data aggregation unit and period in the correlation analysis between temperature and incidence of HFRS in mainland China: A systematic review of 27 ecological studies. PloS Negl. Trop. Dis. 13 (8), e0007688. 10.1371/journal.pntd.0007688 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

