Transcription Factor-Based Biosensor for Dynamic Control in Yeast for Natural Product Synthesis
- PMID: 33614618
- PMCID: PMC7892902
- DOI: 10.3389/fbioe.2021.635265
Transcription Factor-Based Biosensor for Dynamic Control in Yeast for Natural Product Synthesis
Abstract
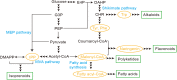
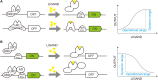
The synthesis of natural products in yeast has gained remarkable achievements with intensive metabolic engineering efforts. In particular, transcription factor (TF)-based biosensors for dynamic control of gene circuits could facilitate strain evaluation, high-throughput screening (HTS), and adaptive laboratory evolution (ALE) for natural product synthesis. In this review, we summarized recent developments of several TF-based biosensors for core intermediates in natural product synthesis through three important pathways, i.e., fatty acid synthesis pathway, shikimate pathway, and methylerythritol-4-phosphate (MEP)/mevalonate (MVA) pathway. Moreover, we have shown how these biosensors are implemented in synthetic circuits for dynamic control of natural product synthesis and also discussed the design/evaluation principles for improved biosensor performance.
Keywords: MEP/MVA pathway; biosensor; fatty acids; natural products; shikimate pathway; transcription factor; yeast.
Copyright © 2021 Zhang and Shi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Transcription factor-based biosensors in biotechnology: current state and future prospects.Appl Microbiol Biotechnol. 2016 Jan;100(1):79-90. doi: 10.1007/s00253-015-7090-3. Epub 2015 Oct 31. Appl Microbiol Biotechnol. 2016. PMID: 26521244 Free PMC article. Review.
-
Biosensors design in yeast and applications in metabolic engineering.FEMS Yeast Res. 2019 Dec 1;19(8):foz082. doi: 10.1093/femsyr/foz082. FEMS Yeast Res. 2019. PMID: 31778177 Review.
-
Synergy between methylerythritol phosphate pathway and mevalonate pathway for isoprene production in Escherichia coli.Metab Eng. 2016 Sep;37:79-91. doi: 10.1016/j.ymben.2016.05.003. Epub 2016 May 9. Metab Eng. 2016. PMID: 27174717
-
Transcription Factor Engineering for High-Throughput Strain Evolution and Organic Acid Bioproduction: A Review.Front Bioeng Biotechnol. 2020 Feb 19;8:98. doi: 10.3389/fbioe.2020.00098. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32140463 Free PMC article. Review.
-
Biosynthesis of β-carotene in engineered E. coli using the MEP and MVA pathways.Microb Cell Fact. 2014 Nov 18;13:160. doi: 10.1186/s12934-014-0160-x. Microb Cell Fact. 2014. PMID: 25403509 Free PMC article.
Cited by
-
A p-Coumaroyl-CoA Biosensor for Dynamic Regulation of Naringenin Biosynthesis in Saccharomyces cerevisiae.ACS Synth Biol. 2022 Oct 21;11(10):3228-3238. doi: 10.1021/acssynbio.2c00111. Epub 2022 Sep 22. ACS Synth Biol. 2022. PMID: 36137537 Free PMC article.
-
Advances in the dynamic control of metabolic pathways in Saccharomyces cerevisiae.Eng Microbiol. 2023 Jun 21;3(4):100103. doi: 10.1016/j.engmic.2023.100103. eCollection 2023 Dec. Eng Microbiol. 2023. PMID: 39628908 Free PMC article. Review.
-
Engineering for life in toxicity: Key to industrializing microbial synthesis of high energy density fuels.Eng Microbiol. 2022 Mar 17;2(2):100013. doi: 10.1016/j.engmic.2022.100013. eCollection 2022 Jun. Eng Microbiol. 2022. PMID: 39628844 Free PMC article. Review.
-
Microbial Transcription Factor-Based Biosensors: Innovations from Design to Applications in Synthetic Biology.Biosensors (Basel). 2025 Mar 31;15(4):221. doi: 10.3390/bios15040221. Biosensors (Basel). 2025. PMID: 40277535 Free PMC article. Review.
-
Directed Evolution of 4-Hydroxyphenylpyruvate Biosensors Based on a Dual Selection System.Int J Mol Sci. 2024 Jan 26;25(3):1533. doi: 10.3390/ijms25031533. Int J Mol Sci. 2024. PMID: 38338812 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

