Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies
- PMID: 33614648
- PMCID: PMC7894079
- DOI: 10.3389/fcell.2021.617281
Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies
Abstract
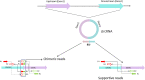
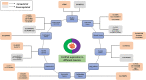
Circular RNAs (circRNAs) are an evolutionarily conserved novel class of non-coding endogenous RNAs (ncRNAs) found in the eukaryotic transcriptome, originally believed to be aberrant RNA splicing by-products with decreased functionality. However, recent advances in high-throughput genomic technology have allowed circRNAs to be characterized in detail and revealed their role in controlling various biological and molecular processes, the most essential being gene regulation. Because of the structural stability, high expression, availability of microRNA (miRNA) binding sites and tissue-specific expression, circRNAs have become hot topic of research in RNA biology. Compared to the linear RNA, circRNAs are produced differentially by backsplicing exons or lariat introns from a pre-messenger RNA (mRNA) forming a covalently closed loop structure missing 3' poly-(A) tail or 5' cap, rendering them immune to exonuclease-mediated degradation. Emerging research has identified multifaceted roles of circRNAs as miRNA and RNA binding protein (RBP) sponges and transcription, translation, and splicing event regulators. CircRNAs have been involved in many human illnesses, including cancer and neurodegenerative disorders such as Alzheimer's and Parkinson's disease, due to their aberrant expression in different pathological conditions. The functional versatility exhibited by circRNAs enables them to serve as potential diagnostic or predictive biomarkers for various diseases. This review discusses the properties, characterization, profiling, and the diverse molecular mechanisms of circRNAs and their use as potential therapeutic targets in different human malignancies.
Keywords: RNA binding protein; circRNA; drug resistance; miRNA sponges; signaling pathways; tumor.
Copyright © 2021 Nisar, Bhat, Singh, Karedath, Rizwan, Hashem, Bagga, Reddy, Jamal, Uddin, Chand, Bedognetti, El-Rifai, Frenneaux, Macha, Ahmed and Haris.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Circular RNAs: A novel type of biomarker and genetic tools in cancer.Oncotarget. 2017 Jun 2;8(38):64551-64563. doi: 10.18632/oncotarget.18350. eCollection 2017 Sep 8. Oncotarget. 2017. PMID: 28969093 Free PMC article. Review.
-
Circular RNAs: Biogenesis, Function and Role in Human Diseases.Front Mol Biosci. 2017 Jun 6;4:38. doi: 10.3389/fmolb.2017.00038. eCollection 2017. Front Mol Biosci. 2017. PMID: 28634583 Free PMC article. Review.
-
Review on circular RNAs and new insights into their roles in cancer.Comput Struct Biotechnol J. 2021 Jan 22;19:910-928. doi: 10.1016/j.csbj.2021.01.018. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33598105 Free PMC article. Review.
-
The emerging role of circRNAs and their clinical significance in human cancers.Biochim Biophys Acta Rev Cancer. 2018 Dec;1870(2):247-260. doi: 10.1016/j.bbcan.2018.06.002. Epub 2018 Jun 19. Biochim Biophys Acta Rev Cancer. 2018. PMID: 29928954 Review.
-
The emerging landscape of circular RNA in life processes.RNA Biol. 2017 Aug 3;14(8):992-999. doi: 10.1080/15476286.2016.1220473. Epub 2016 Aug 11. RNA Biol. 2017. PMID: 27617908 Free PMC article. Review.
Cited by
-
Circular RNAs: a small piece in the heart failure puzzle.Funct Integr Genomics. 2024 May 17;24(3):102. doi: 10.1007/s10142-024-01386-z. Funct Integr Genomics. 2024. PMID: 38760573 Review.
-
circGlis3 promotes β-cell dysfunction by binding to heterogeneous nuclear ribonucleoprotein F and encoding Glis3-348aa protein.iScience. 2023 Dec 9;27(1):108680. doi: 10.1016/j.isci.2023.108680. eCollection 2024 Jan 19. iScience. 2023. PMID: 38226164 Free PMC article.
-
Non-Coding RNAs Regulating Mitochondrial Functions and the Oxidative Stress Response as Putative Targets against Age-Related Macular Degeneration (AMD).Int J Mol Sci. 2023 Jan 30;24(3):2636. doi: 10.3390/ijms24032636. Int J Mol Sci. 2023. PMID: 36768958 Free PMC article. Review.
-
Ferroptosis-associated circular RNAs: Opportunities and challenges in the diagnosis and treatment of cancer.Front Cell Dev Biol. 2023 Apr 20;11:1160381. doi: 10.3389/fcell.2023.1160381. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37152286 Free PMC article. Review.
-
Circular RNA hsa_circ_0001874 is an indicator for gastric cancer.J Clin Lab Anal. 2021 Jul;35(7):e23851. doi: 10.1002/jcla.23851. Epub 2021 May 24. J Clin Lab Anal. 2021. PMID: 34028890 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

