Targeting Bromodomain and Extraterminal Proteins for Drug Discovery: From Current Progress to Technological Development
- PMID: 33616410
- PMCID: PMC8106541
- DOI: 10.1021/acs.jmedchem.0c01487
Targeting Bromodomain and Extraterminal Proteins for Drug Discovery: From Current Progress to Technological Development
Abstract
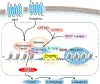
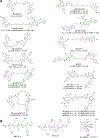
Bromodomain and extraterminal (BET) proteins bind acetylated lysine residues in histones and nonhistone proteins via tandem bromodomains and regulate chromatin dynamics, cellular processes, and disease procession. Thus targeting BET proteins is a promising strategy for treating various diseases, especially malignant tumors and chronic inflammation. Many pan-BET small-molecule inhibitors have been described, and some of them are in clinical evaluation. Nevertheless, the limited clinical efficacy of the current BET inhibitors is also evident and has inspired the development of new technologies to improve their clinical outcomes and minimize unwanted side effects. In this Review, we summarize the latest protein characteristics and biological functions of BRD4 as an example of BET proteins, analyze the clinical development status and preclinical resistance mechanisms, and discuss recent advances in BRD4-selective inhibitors, dual-target BET inhibitors, proteolysis targeting chimera degraders, and protein-protein interaction inhibitors.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Covalent-Fragment Screening of BRD4 Identifies a Ligandable Site Orthogonal to the Acetyl-Lysine Binding Sites.ACS Chem Biol. 2020 Apr 17;15(4):1036-1049. doi: 10.1021/acschembio.0c00058. Epub 2020 Mar 23. ACS Chem Biol. 2020. PMID: 32149490 Free PMC article.
-
Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.Nature. 2020 Feb;578(7794):306-310. doi: 10.1038/s41586-020-1930-8. Epub 2020 Jan 22. Nature. 2020. PMID: 31969702
-
BET Bromodomain as a Target of Epigenetic Therapy.Chem Pharm Bull (Tokyo). 2016;64(6):540-7. doi: 10.1248/cpb.c16-00225. Chem Pharm Bull (Tokyo). 2016. PMID: 27250788 Review.
-
Targeting the epigenetic reader "BET" as a therapeutic strategy for cancer.Bioorg Chem. 2023 Nov;140:106833. doi: 10.1016/j.bioorg.2023.106833. Epub 2023 Sep 4. Bioorg Chem. 2023. PMID: 37683545 Review.
-
Targeting Bromodomain-Selective Inhibitors of BET Proteins in Drug Discovery and Development.J Med Chem. 2022 Apr 14;65(7):5184-5211. doi: 10.1021/acs.jmedchem.1c01835. Epub 2022 Mar 24. J Med Chem. 2022. PMID: 35324195
Cited by
-
Application of PROTACs in Target Identification and Target Validation.Acta Mater Med. 2024 Feb 21;3(1):72-87. doi: 10.15212/amm-2024-0010. Epub 2024 Mar 21. Acta Mater Med. 2024. PMID: 39373008 Free PMC article.
-
Medicinal Chemistry of Anti-HIV-1 Latency Chemotherapeutics: Biotargets, Binding Modes and Structure-Activity Relationship Investigation.Molecules. 2022 Dec 20;28(1):3. doi: 10.3390/molecules28010003. Molecules. 2022. PMID: 36615199 Free PMC article. Review.
-
IDR-targeting compounds suppress HPV genome replication via disruption of phospho-BRD4 association with DNA damage response factors.Mol Cell. 2024 Jan 18;84(2):202-220.e15. doi: 10.1016/j.molcel.2023.11.022. Epub 2023 Dec 15. Mol Cell. 2024. PMID: 38103559 Free PMC article.
-
Identification of a SNAI1 enhancer RNA that drives cancer cell plasticity.Nat Commun. 2025 Mar 25;16(1):2890. doi: 10.1038/s41467-025-58032-w. Nat Commun. 2025. PMID: 40133308 Free PMC article.
-
An updated patent review of BRD4 degraders.Expert Opin Ther Pat. 2024 Oct;34(10):929-951. doi: 10.1080/13543776.2024.2400166. Epub 2024 Sep 4. Expert Opin Ther Pat. 2024. PMID: 39219068 Review.
References
-
- Wu SY; Chiang CM The double bromodomain-containing chromatin adaptor BRD4 and transcriptional regulation. J. Biol. Chem 2007, 282, 13141–13145. - PubMed
-
- Dhalluin C; Carlson JE; Zeng L; He C; Aggarwal AK; Zhou M-M; Zhou M-M Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999, 399, 491–496. - PubMed
-
- Filippakopoulos P; Knapp S Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discovery 2014, 13, 337–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases

