Megasphaera lornae sp. nov., Megasphaera hutchinsoni sp. nov., and Megasphaera vaginalis sp. nov.: novel bacteria isolated from the female genital tract
- PMID: 33616513
- PMCID: PMC8161547
- DOI: 10.1099/ijsem.0.004702
Megasphaera lornae sp. nov., Megasphaera hutchinsoni sp. nov., and Megasphaera vaginalis sp. nov.: novel bacteria isolated from the female genital tract
Abstract
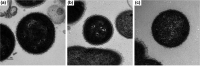
Six strictly anaerobic Gram-negative bacteria representing three novel species were isolated from the female reproductive tract. The proposed type strains for each species were designated UPII 199-6T, KA00182T and BV3C16-1T. Phylogenetic analyses based on 16S rRNA gene sequencing indicated that the bacterial isolates were members of the genus Megasphaera. UPII 199-6T and KA00182T had 16S rRNA gene sequence identities of 99.9 % with 16S rRNA clone sequences previously amplified from the human vagina designated as Megasphaera type 1 and Megasphaera type 2, members of the human vaginal microbiota associated with bacterial vaginosis, preterm birth and HIV acquisition. UPII 199-6T exhibited sequence identities ranging from 92.9 to 93.6 % with validly named Megasphaera isolates and KA00182T had 16S rRNA gene sequence identities ranging from 92.6-94.2 %. BV3C16-1T was most closely related to Megasphaera cerevisiae with a 16S rRNA gene sequence identity of 95.4 %. Cells were coccoid or diplococcoid, non-motile and did not form spores. Genital tract isolates metabolized organic acids but were asaccharolytic. The isolates also metabolized amino acids. The DNA G+C content for the genome sequences of UPII 199-6T, KA00182T and BV3C16-1T were 46.4, 38.9 and 49.8 mol%, respectively. Digital DNA-DNA hybridization and average nucleotide identity between the genital tract isolates and other validly named Megasphaera species suggest that each isolate type represents a new species. The major fatty acid methyl esters include the following: C12 : 0, C16 : 0, C16 : 0 dimethyl acetal (DMA) and summed feature 5 (C15 : 0 DMA and/or C14 : 0 3-OH) in UPII 199-6T; C16 : 0 and C16 : 1 cis 9 in KA00182T; C12 : 0; C14 : 0 3-OH; and summed feature 5 in BV3C16-1T. The isolates produced butyrate, isobutyrate, and isovalerate but there were specific differences including production of formate and propionate. Together, these data indicate that UPII 199-6T, KA00182T and BV3C16-1T represent novel species within the genus Megasphaera. We propose the following names: Megasphaera lornae sp. nov. for UPII 199-6T representing the type strain of this species (=DSM 111201T=ATCC TSD-205T), Megasphaera hutchinsoni sp. nov. for KA00182T representing the type strain of this species (=DSM 111202T=ATCC TSD-206T) and Megasphaera vaginalis sp. nov. for BV3C16-1T representing the type strain of this species (=DSM 111203T=ATCC TSD-207T).
Keywords: Megasphaera; Negativicutes; Veillonellaceae; bacterial vaginosis; genital tract bacteria; human vagina.
Conflict of interest statement
D. N. F. and T. F., have received a royalty from B. D., around molecular diagnosis of B. V. S. S., has received speaking honoraria from Lupin Inc. S. L. H., has served as a consultant to Hologic related to the development of diagnostic tests for bacterial vaginosis and her institution has received research funding from Cepheid and Becton-Dickinson.
Figures



References
-
- Rogosa M. Transfer of Peptostreptococcus elsdenii to a new genus, Megasphaera [M. elsdenii Gutierrez, et al. comb. nov.]. Int J Syst Bact. 1971;21:187–189. doi: 10.1099/00207713-21-2-187. - DOI
-
- Engelmann U, Weiss N. Megasphaera cerevisiae sp. nov.: a new Gram-negative obligately anaerobic coccus isolated from spoiled beer. Syst Appl Microbiol. 1985;6:287–290. doi: 10.1016/S0723-2020(85)80033-3. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

