Mitonuclear Coevolution, but not Nuclear Compensation, Drives Evolution of OXPHOS Complexes in Bivalves
- PMID: 33616640
- PMCID: PMC8136519
- DOI: 10.1093/molbev/msab054
Mitonuclear Coevolution, but not Nuclear Compensation, Drives Evolution of OXPHOS Complexes in Bivalves
Abstract
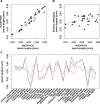
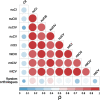
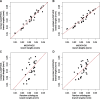
In Metazoa, four out of five complexes involved in oxidative phosphorylation (OXPHOS) are formed by subunits encoded by both the mitochondrial (mtDNA) and nuclear (nuDNA) genomes, leading to the expectation of mitonuclear coevolution. Previous studies have supported coadaptation of mitochondria-encoded (mtOXPHOS) and nuclear-encoded OXPHOS (nuOXPHOS) subunits, often specifically interpreted with regard to the "nuclear compensation hypothesis," a specific form of mitonuclear coevolution where nuclear genes compensate for deleterious mitochondrial mutations due to less efficient mitochondrial selection. In this study, we analyzed patterns of sequence evolution of 79 OXPHOS subunits in 31 bivalve species, a taxon showing extraordinary mtDNA variability and including species with "doubly uniparental" mtDNA inheritance. Our data showed strong and clear signals of mitonuclear coevolution. NuOXPHOS subunits had concordant topologies with mtOXPHOS subunits, contrary to previous phylogenies based on nuclear genes lacking mt interactions. Evolutionary rates between mt and nuOXPHOS subunits were also highly correlated compared with non-OXPHO-interacting nuclear genes. Nuclear subunits of chimeric OXPHOS complexes (I, III, IV, and V) also had higher dN/dS ratios than Complex II, which is formed exclusively by nuDNA-encoded subunits. However, we did not find evidence of nuclear compensation: mitochondria-encoded subunits showed similar dN/dS ratios compared with nuclear-encoded subunits, contrary to most previously studied bilaterian animals. Moreover, no site-specific signals of compensatory positive selection were detected in nuOXPHOS genes. Our analyses extend the evidence for mitonuclear coevolution to a new taxonomic group, but we propose a reconsideration of the nuclear compensation hypothesis.
Keywords: OXPHOS; bivalvia; compensatory evolution; evolutionary rates; mitonuclear coevolution.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Allio R, Donega S, Galtier N, Nabholz B.. 2017. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol Biol Evol. 34(11):2762–2772. - PubMed
-
- Anisimova M, Bielawski JP, Yang Z.. 2002. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 19(6):950–958. - PubMed
-
- Ballard JW, Whitlock MC.. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13(4):729–744. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

