Genomic Epidemiology Analysis of Infectious Disease Outbreaks Using TransPhylo
- PMID: 33617114
- PMCID: PMC7995038
- DOI: 10.1002/cpz1.60
Genomic Epidemiology Analysis of Infectious Disease Outbreaks Using TransPhylo
Erratum in
-
Group Correction Statement (Data Availability Statements).Curr Protoc. 2022 Aug;2(8):e552. doi: 10.1002/cpz1.552. Curr Protoc. 2022. PMID: 36005902 Free PMC article. No abstract available.
-
Group Correction Statement (Conflict of Interest Statements).Curr Protoc. 2022 Aug;2(8):e551. doi: 10.1002/cpz1.551. Curr Protoc. 2022. PMID: 36005903 Free PMC article. No abstract available.
Abstract
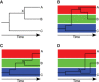
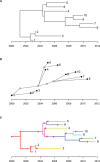
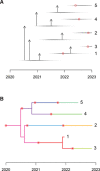
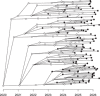
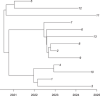
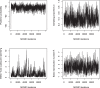
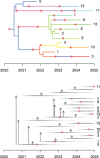
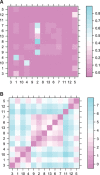
Comparing the pathogen genomes from several cases of an infectious disease has the potential to help us understand and control outbreaks. Many methods exist to reconstruct a phylogeny from such genomes, which represents how the genomes are related to one another. However, such a phylogeny is not directly informative about transmission events between individuals. TransPhylo is a software tool implemented as an R package designed to bridge the gap between pathogen phylogenies and transmission trees. TransPhylo is based on a combined model of transmission between hosts and pathogen evolution within each host. It can simulate both phylogenies and transmission trees jointly under this combined model. TransPhylo can also reconstruct a transmission tree based on a dated phylogeny, by exploring the space of transmission trees compatible with the phylogeny. A transmission tree can be represented as a coloring of a phylogeny where each color represents a different host of the pathogen, and TransPhylo provides convenient ways to plot these colorings and explore the results. This article presents the basic protocols that can be used to make the most of TransPhylo. © 2021 The Authors. Basic Protocol 1: First steps with TransPhylo Basic Protocol 2: Simulation of outbreak data Basic Protocol 3: Inference of transmission Basic Protocol 4: Exploring the results of inference.
Keywords: genomic epidemiology; infectious disease outbreak; phylogenetics; transmission analysis.
© 2021 The Authors.
Figures
Similar articles
-
Genomic Infectious Disease Epidemiology in Partially Sampled and Ongoing Outbreaks.Mol Biol Evol. 2017 Apr 1;34(4):997-1007. doi: 10.1093/molbev/msw275. Mol Biol Evol. 2017. PMID: 28100788 Free PMC article.
-
Inference of Infectious Disease Transmission through a Relaxed Bottleneck Using Multiple Genomes Per Host.Mol Biol Evol. 2024 Jan 3;41(1):msad288. doi: 10.1093/molbev/msad288. Mol Biol Evol. 2024. PMID: 38168711 Free PMC article.
-
Incorporating Epidemiological Data into the Genomic Analysis of Partially Sampled Infectious Disease Outbreaks.Mol Biol Evol. 2025 Apr 1;42(4):msaf083. doi: 10.1093/molbev/msaf083. Mol Biol Evol. 2025. PMID: 40256930 Free PMC article.
-
The role of pathogen genomics in assessing disease transmission.BMJ. 2015 May 11;350:h1314. doi: 10.1136/bmj.h1314. BMJ. 2015. PMID: 25964672 Review.
-
Microbial Genomics of Ancient Plagues and Outbreaks.Trends Microbiol. 2016 Dec;24(12):978-990. doi: 10.1016/j.tim.2016.08.004. Epub 2016 Sep 8. Trends Microbiol. 2016. PMID: 27618404 Review.
Cited by
-
Modeling approaches for early warning and monitoring of pandemic situations as well as decision support.Front Public Health. 2022 Nov 14;10:994949. doi: 10.3389/fpubh.2022.994949. eCollection 2022. Front Public Health. 2022. PMID: 36452960 Free PMC article. Review.
-
Clinical and Molecular Characterizations of Carbapenem-Resistant Klebsiella pneumoniae Causing Bloodstream Infection in a Chinese Hospital.Microbiol Spectr. 2022 Oct 26;10(5):e0169022. doi: 10.1128/spectrum.01690-22. Epub 2022 Oct 3. Microbiol Spectr. 2022. PMID: 36190403 Free PMC article.
-
Transmission history of SARS-CoV-2 in humans and white-tailed deer.Sci Rep. 2022 Jul 15;12(1):12094. doi: 10.1038/s41598-022-16071-z. Sci Rep. 2022. PMID: 35840592 Free PMC article.
-
The NOSTRA model: Coherent estimation of infection sources in the case of possible nosocomial transmission.PLoS Comput Biol. 2025 Apr 21;21(4):e1012949. doi: 10.1371/journal.pcbi.1012949. eCollection 2025 Apr. PLoS Comput Biol. 2025. PMID: 40258227 Free PMC article.
-
Apollo: a comprehensive GPU-powered within-host simulator for viral evolution and infection dynamics across population, tissue, and cell.Nat Commun. 2025 Jul 1;16(1):5783. doi: 10.1038/s41467-025-60988-8. Nat Commun. 2025. PMID: 40593638 Free PMC article.
References
Literature Cited
-
- Ayabina, D. , Ronning, J. O. , Alfsnes, K. , Debech, N. , Brynildsrud, O. B. , Arnesen, T. , … Eldholm, V. (2018). Genome‐based transmission modelling separates imported tuberculosis from recent transmission within an immigrant population. Microbial Genomics, 4, e000219. doi: 10.1099/mgen.0.000219. - DOI - PMC - PubMed
-
- Cheng, V. C. C. , Wong, S. C. , Cao, H. , Chen, J. H. K. , So, S. Y. C. , Wong, S. C. Y. , … Ho, P. L. (2019). Whole‐genome sequencing data‐based modeling for the investigation of an outbreak of community‐associated methicillin‐resistant Staphylococcus aureus in a neonatal intensive care unit in Hong Kong. European Journal of Clinical Microbiology and Infectious Diseases, 38, 563–573. doi: 10.1007/s10096-018-03458-y. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

