Selective targeting of the inactive state of hematopoietic cell kinase (Hck) with a stable curcumin derivative
- PMID: 33617879
- PMCID: PMC7946438
- DOI: 10.1016/j.jbc.2021.100449
Selective targeting of the inactive state of hematopoietic cell kinase (Hck) with a stable curcumin derivative
Abstract
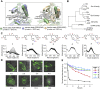
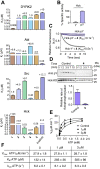
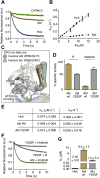
Hck, a Src family nonreceptor tyrosine kinase (SFK), has recently been established as an attractive pharmacological target to improve pulmonary function in COVID-19 patients. Hck inhibitors are also well known for their regulatory role in various malignancies and autoimmune diseases. Curcumin has been previously identified as an excellent DYRK-2 inhibitor, but curcumin's fate is tainted by its instability in the cellular environment. Besides, small molecules targeting the inactive states of a kinase are desirable to reduce promiscuity. Here, we show that functionalization of the 4-arylidene position of the fluorescent curcumin scaffold with an aryl nitrogen mustard provides a stable Hck inhibitor (Kd = 50 ± 10 nM). The mustard curcumin derivative preferentially interacts with the inactive conformation of Hck, similar to type-II kinase inhibitors that are less promiscuous. Moreover, the lead compound showed no inhibitory effect on three other kinases (DYRK2, Src, and Abl). We demonstrate that the cytotoxicity may be mediated via inhibition of the SFK signaling pathway in triple-negative breast cancer and murine macrophage cells. Our data suggest that curcumin is a modifiable fluorescent scaffold to develop selective kinase inhibitors by remodeling its target affinity and cellular stability.
Keywords: Hck inhibitor; Src family kinase; cell signaling; curcumin derivative; enzyme kinetics; kinase inhibition.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
Constitutive activation of the Src-family kinases Fgr and Hck enhances the tumor burden of acute myeloid leukemia cells in immunocompromised mice.Sci Rep. 2025 Jan 2;15(1):174. doi: 10.1038/s41598-024-83740-6. Sci Rep. 2025. PMID: 39747387 Free PMC article.
-
Interaction with the Src homology (SH3-SH2) region of the Src-family kinase Hck structures the HIV-1 Nef dimer for kinase activation and effector recruitment.J Biol Chem. 2014 Oct 10;289(41):28539-53. doi: 10.1074/jbc.M114.600031. Epub 2014 Aug 13. J Biol Chem. 2014. PMID: 25122770 Free PMC article.
-
Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.Acta Crystallogr D Biol Crystallogr. 2014 Feb;70(Pt 2):392-404. doi: 10.1107/S1399004713028654. Epub 2014 Jan 29. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24531473
-
Hck inhibitors as potential therapeutic agents in cancer and HIV infection.Curr Med Chem. 2015;22(13):1540-64. doi: 10.2174/0929867322666150209152057. Curr Med Chem. 2015. PMID: 25666803 Review.
-
Insights on hematopoietic cell kinase: An oncogenic player in human cancer.Biomed Pharmacother. 2023 Apr;160:114339. doi: 10.1016/j.biopha.2023.114339. Epub 2023 Feb 1. Biomed Pharmacother. 2023. PMID: 36736283 Review.
Cited by
-
Bioinformatics analysis of potential pathogenesis and risk genes of immunoinflammation-promoted renal injury in severe COVID-19.Front Immunol. 2022 Aug 16;13:950076. doi: 10.3389/fimmu.2022.950076. eCollection 2022. Front Immunol. 2022. PMID: 36052061 Free PMC article.
-
Unraveling the mechanism of action of cepharanthine for the treatment of novel coronavirus pneumonia (COVID-19) from the perspectives of systematic pharmacology.Arab J Chem. 2023 Jun;16(6):104722. doi: 10.1016/j.arabjc.2023.104722. Epub 2023 Mar 6. Arab J Chem. 2023. PMID: 36910427 Free PMC article.
References
-
- Knight Z.A., Shokat K.M. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. - PubMed
-
- Li Y., Zou Q., Yuan C., Li S., Xing R., Yan X. Amino acid coordination driven self-assembly for enhancing both the biological stability and tumor accumulation of curcumin. Angew. Chem. Int. Ed. Engl. 2018;57:17084–17088. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

