Binding Dynamics of Disordered Linker Histone H1 with a Nucleosomal Particle
- PMID: 33617899
- PMCID: PMC9272445
- DOI: 10.1016/j.jmb.2021.166881
Binding Dynamics of Disordered Linker Histone H1 with a Nucleosomal Particle
Abstract
Linker histone H1 is an essential regulatory protein for many critical biological processes, such as eukaryotic chromatin packaging and gene expression. Mis-regulation of H1s is commonly observed in tumor cells, where the balance between different H1 subtypes has been shown to alter the cancer phenotype. Consisting of a rigid globular domain and two highly charged terminal domains, H1 can bind to multiple sites on a nucleosomal particle to alter chromatin hierarchical condensation levels. In particular, the disordered H1 amino- and carboxyl-terminal domains (NTD/CTD) are believed to enhance this binding affinity, but their detailed dynamics and functions remain unclear. In this work, we used a coarse-grained computational model, AWSEM-DNA, to simulate the H1.0b-nucleosome complex, namely chromatosome. Our results demonstrate that H1 disordered domains restrict the dynamics and conformation of both globular H1 and linker DNA arms, resulting in a more compact and rigid chromatosome particle. Furthermore, we identified regions of H1 disordered domains that are tightly tethered to DNA near the entry-exit site. Overall, our study elucidates at near-atomic resolution the way the disordered linker histone H1 modulates nucleosome's structural preferences and conformational dynamics.
Keywords: IDP; MD simulation; chromatin; chromatosome; protein-DNA interaction.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
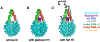

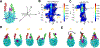

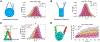
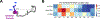
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

