Tricks and threats of RNA viruses - towards understanding the fate of viral RNA
- PMID: 33618611
- PMCID: PMC8078519
- DOI: 10.1080/15476286.2021.1875680
Tricks and threats of RNA viruses - towards understanding the fate of viral RNA
Abstract
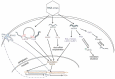
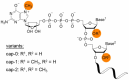
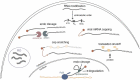
Human innate cellular defence pathways have evolved to sense and eliminate pathogens, of which, viruses are considered one of the most dangerous. Their relatively simple structure makes the identification of viral invasion a difficult task for cells. In the course of evolution, viral nucleic acids have become one of the strongest and most reliable early identifiers of infection. When considering RNA virus recognition, RNA sensing is the central mechanism in human innate immunity, and effectiveness of this sensing is crucial for triggering an appropriate antiviral response. Although human cells are armed with a variety of highly specialized receptors designed to respond only to pathogenic viral RNA, RNA viruses have developed an array of mechanisms to avoid being recognized by human interferon-mediated cellular defence systems. The repertoire of viral evasion strategies is extremely wide, ranging from masking pathogenic RNA through end modification, to utilizing sophisticated techniques to deceive host cellular RNA degrading enzymes, and hijacking the most basic metabolic pathways in host cells. In this review, we aim to dissect human RNA sensing mechanisms crucial for antiviral immune defences, as well as the strategies adopted by RNA viruses to avoid detection and degradation by host cells. We believe that understanding the fate of viral RNA upon infection, and detailing the molecular mechanisms behind virus-host interactions, may be helpful for developing more effective antiviral strategies; which are urgently needed to prevent the far-reaching consequences of widespread, highly pathogenic viral infections.
Keywords: RNA viruses; viral RNA degradation; viral RNA sensing; viral evasion.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
Crosstalk between nucleocytoplasmic trafficking and the innate immune response to viral infection.J Biol Chem. 2021 Jul;297(1):100856. doi: 10.1016/j.jbc.2021.100856. Epub 2021 Jun 29. J Biol Chem. 2021. PMID: 34097873 Free PMC article. Review.
-
Slicing and dicing viruses: antiviral RNA interference in mammals.EMBO J. 2019 Apr 15;38(8):e100941. doi: 10.15252/embj.2018100941. Epub 2019 Mar 14. EMBO J. 2019. PMID: 30872283 Free PMC article. Review.
-
Roadblocks and fast tracks: How RNA binding proteins affect the viral RNA journey in the cell.Semin Cell Dev Biol. 2021 Mar;111:86-100. doi: 10.1016/j.semcdb.2020.08.006. Epub 2020 Aug 23. Semin Cell Dev Biol. 2021. PMID: 32847707 Free PMC article. Review.
-
Interferon and immunity: the role of microRNA in viral evasion strategies.Front Immunol. 2025 May 9;16:1567459. doi: 10.3389/fimmu.2025.1567459. eCollection 2025. Front Immunol. 2025. PMID: 40416980 Free PMC article. Review.
-
Mechanisms of innate immune evasion in re-emerging RNA viruses.Curr Opin Virol. 2015 Jun;12:26-37. doi: 10.1016/j.coviro.2015.02.005. Epub 2015 Mar 9. Curr Opin Virol. 2015. PMID: 25765605 Free PMC article. Review.
Cited by
-
Effective recognition of double-stranded RNA does not require activation of cellular inflammation.Sci Adv. 2025 Apr 11;11(15):eads6498. doi: 10.1126/sciadv.ads6498. Epub 2025 Apr 9. Sci Adv. 2025. PMID: 40203104 Free PMC article.
-
Trypanosome mRNA recapping is triggered by hypermethylation originating from cap 4.Nucleic Acids Res. 2024 Sep 23;52(17):10645-10653. doi: 10.1093/nar/gkae614. Nucleic Acids Res. 2024. PMID: 39011881 Free PMC article.
-
Placenta - A Competent, But Not Infallible, Antiviral and Antiparasitic Barrier.Reprod Sci. 2025 Aug;32(8):2669-2684. doi: 10.1007/s43032-025-01921-8. Epub 2025 Jul 14. Reprod Sci. 2025. PMID: 40660015 Review. No abstract available.
-
Natural and Designed Cyclic Peptides as Potential Antiviral Drugs to Combat Future Coronavirus Outbreaks.Molecules. 2025 Apr 8;30(8):1651. doi: 10.3390/molecules30081651. Molecules. 2025. PMID: 40333520 Free PMC article. Review.
-
Molecular Mechanisms Underlying Response to Influenza in Grey Seals (Halichoerus grypus), a Potential Wild Reservoir.Mol Ecol. 2025 Aug;34(15):e70012. doi: 10.1111/mec.70012. Epub 2025 Jul 4. Mol Ecol. 2025. PMID: 40613337 Free PMC article.
References
-
- Pichlmair A, Lassnig C, Eberle C-A, et al. IFIT1 Is an Antiviral Protein That Recognizes 5′-Triphosphate RNA. Nat Immunol. 2011;12(7):624–630. - PubMed
-
- Platnich JM, Muruve DA. NOD-like Receptors and Inflammasomes: A Review of Their Canonical and Non-Canonical Signaling Pathways. Arch Biochem Biophys. 2019;670:4–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
