L-plastin Ser5 phosphorylation is modulated by the PI3K/SGK pathway and promotes breast cancer cell invasiveness
- PMID: 33618712
- PMCID: PMC7898450
- DOI: 10.1186/s12964-021-00710-5
L-plastin Ser5 phosphorylation is modulated by the PI3K/SGK pathway and promotes breast cancer cell invasiveness
Abstract
Background: Metastasis is the predominant cause for cancer morbidity and mortality accounting for approximatively 90% of cancer deaths. The actin-bundling protein L-plastin has been proposed as a metastatic marker and phosphorylation on its residue Ser5 is known to increase its actin-bundling activity. We recently showed that activation of the ERK/MAPK signalling pathway leads to L-plastin Ser5 phosphorylation and that the downstream kinases RSK1 and RSK2 are able to directly phosphorylate Ser5. Here we investigate the involvement of the PI3K pathway in L-plastin Ser5 phosphorylation and the functional effect of this phosphorylation event in breast cancer cells.
Methods: To unravel the signal transduction network upstream of L-plastin Ser5 phosphorylation, we performed computational modelling based on immunoblot analysis data, followed by experimental validation through inhibition/overexpression studies and in vitro kinase assays. To assess the functional impact of L-plastin expression/Ser5 phosphorylation in breast cancer cells, we either silenced L-plastin in cell lines initially expressing endogenous L-plastin or neoexpressed L-plastin wild type and phosphovariants in cell lines devoid of endogenous L-plastin. The established cell lines were used for cell biology experiments and confocal microscopy analysis.
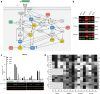
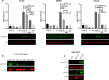
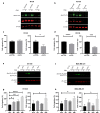
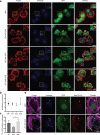
Results: Our modelling approach revealed that, in addition to the ERK/MAPK pathway and depending on the cellular context, the PI3K pathway contributes to L-plastin Ser5 phosphorylation through its downstream kinase SGK3. The results of the transwell invasion/migration assays showed that shRNA-mediated knockdown of L-plastin in BT-20 or HCC38 cells significantly reduced cell invasion, whereas stable expression of the phosphomimetic L-plastin Ser5Glu variant led to increased migration and invasion of BT-549 and MDA-MB-231 cells. Finally, confocal image analysis combined with zymography experiments and gelatin degradation assays provided evidence that L-plastin Ser5 phosphorylation promotes L-plastin recruitment to invadopodia, MMP-9 activity and concomitant extracellular matrix degradation.
Conclusion: Altogether, our results demonstrate that L-plastin Ser5 phosphorylation increases breast cancer cell invasiveness. Being a downstream molecule of both ERK/MAPK and PI3K/SGK pathways, L-plastin is proposed here as a potential target for therapeutic approaches that are aimed at blocking dysregulated signalling outcome of both pathways and, thus, at impairing cancer cell invasion and metastasis formation. Video abstract.
Keywords: Actin-bundling; ERK/MAPK pathway; Extracellular matrix degradation; Invadopodia; Invasion; L-plastin; Metastasis; PI3K pathway; RSK; SGK.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
L-plastin Ser5 phosphorylation in breast cancer cells and in vitro is mediated by RSK downstream of the ERK/MAPK pathway.FASEB J. 2016 Mar;30(3):1218-33. doi: 10.1096/fj.15-276311. Epub 2015 Nov 30. FASEB J. 2016. PMID: 26631483
-
Quantitative kinetic study of the actin-bundling protein L-plastin and of its impact on actin turn-over.PLoS One. 2010 Feb 15;5(2):e9210. doi: 10.1371/journal.pone.0009210. PLoS One. 2010. PMID: 20169155 Free PMC article.
-
Phosphorylation on Ser5 increases the F-actin-binding activity of L-plastin and promotes its targeting to sites of actin assembly in cells.J Cell Sci. 2006 May 1;119(Pt 9):1947-60. doi: 10.1242/jcs.02874. J Cell Sci. 2006. PMID: 16636079
-
The actin-bundling protein L-plastin supports T-cell motility and activation.Immunol Rev. 2013 Nov;256(1):48-62. doi: 10.1111/imr.12102. Immunol Rev. 2013. PMID: 24117812 Free PMC article. Review.
-
L-plastin regulates the stability of the immune synapse of naive and effector T-cells.Adv Biol Regul. 2017 Jan;63:107-114. doi: 10.1016/j.jbior.2016.09.009. Epub 2016 Sep 27. Adv Biol Regul. 2017. PMID: 27720134 Review.
Cited by
-
ZAP-70 augments tonic B-cell receptor and CCR7 signaling in IGHV-unmutated chronic lymphocytic leukemia.Blood Adv. 2024 Mar 12;8(5):1167-1178. doi: 10.1182/bloodadvances.2022009557. Blood Adv. 2024. PMID: 38113463 Free PMC article.
-
T-plastin contributes to epithelial-mesenchymal transition in human lung cancer cells through FAK/AKT/Slug axis signaling pathway.BMB Rep. 2024 Jun;57(6):305-310. doi: 10.5483/BMBRep.2024-0040. BMB Rep. 2024. PMID: 38835117 Free PMC article.
-
TEM1 up-regulates MMP-2 and promotes ECM remodeling for facilitating invasion and migration of uterine sarcoma.Discov Oncol. 2023 Jan 13;14(1):5. doi: 10.1007/s12672-023-00613-6. Discov Oncol. 2023. PMID: 36639546 Free PMC article.
-
Human plastins are novel cytoskeletal pH sensors with a reduced F-actin bundling capacity at basic pH.bioRxiv [Preprint]. 2025 Mar 28:2025.03.26.645573. doi: 10.1101/2025.03.26.645573. bioRxiv. 2025. Update in: J Mol Biol. 2025 Oct 1;437(19):169306. doi: 10.1016/j.jmb.2025.169306. PMID: 40196613 Free PMC article. Updated. Preprint.
-
Human Plastins are Novel Cytoskeletal pH Sensors with a Reduced F-actin Bundling Capacity at Basic pH.J Mol Biol. 2025 Oct 1;437(19):169306. doi: 10.1016/j.jmb.2025.169306. Epub 2025 Jun 25. J Mol Biol. 2025. PMID: 40578670 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

