GS-9822, a preclinical LEDGIN candidate, displays a block-and-lock phenotype in cell culture
- PMID: 33619061
- PMCID: PMC8092873
- DOI: 10.1128/AAC.02328-20
GS-9822, a preclinical LEDGIN candidate, displays a block-and-lock phenotype in cell culture
Abstract
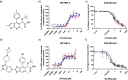
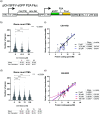
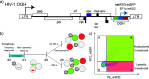
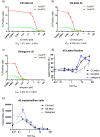
The ability of HIV to integrate into the host genome and establish latent reservoirs is the main hurdle preventing an HIV cure. LEDGINs are small-molecule integrase inhibitors that target the binding pocket of LEDGF/p75, a cellular cofactor that substantially contributes to HIV integration site selection. They are potent antivirals that inhibit HIV integration and maturation. In addition, they retarget residual integrants away from transcription units and towards a more repressive chromatin environment. As a result, treatment with the LEDGIN CX14442 yielded residual provirus that proved more latent and more refractory to reactivation, supporting the use of LEDGINs as research tools to study HIV latency and a functional cure strategy. In this study we compared GS-9822, a potent, pre-clinical lead compound, with CX14442 with respect to antiviral potency, integration site selection, latency and reactivation. GS-9822 was more potent than CX14442 in most assays. For the first time, the combined effects on viral replication, integrase-LEDGF/p75 interaction, integration sites, epigenetic landscape, immediate latency and latency reversal was demonstrated at nanomolar concentrations achievable in the clinic. GS-9822 profiles as a preclinical candidate for future functional cure research.
Copyright © 2021 American Society for Microbiology.
Figures

References
-
- National Institutes of Health. 2021. FDA-approved HIV medicines. Understanding HIV/AIDS. AIDSinfo. NIH, Bethesda, MD. https://hivinfo.nih.gov/understanding-hiv/fact-sheets/fda-approved-hiv-m....
-
- EACS. 2019. European guidelines for the treatment of people living with HIV (PLWH) in Europe. Guidelines 122. EACS, Brussels, Belgium. https://www.eacsociety.org/guidelines/eacs-guidelines/eacs-guidelines.html.
-
- DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents–A Working Group of the Office of AIDS Research Advisory Council (OARAC). 2018. Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in adults and adolescents with HIV. Department of Health and Human Services, Washington, DC. https://clinicalinfo.hiv.gov/sites/default/files/inline-files/AdultandAd....
-
- World Health Organization (WHO). 2019. Policy brief: update of recommendations on first- and second-line antiretroviral regimens. WHO, Geneva, Switzerland. https://www.who.int/hiv/pub/arv/arv-update-2019-policy/en/.
-
- Joint United Nations Programme on HIV/AIDS (UNAIDS). 2019. UNAIDS data 2019. UNAIDS, Geneva, Switzerland. https://www.unaids.org/en/resources/documents/2019/2019-UNAIDS-data.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

