Gatorbulin-1, a distinct cyclodepsipeptide chemotype, targets a seventh tubulin pharmacological site
- PMID: 33619102
- PMCID: PMC7936326
- DOI: 10.1073/pnas.2021847118
Gatorbulin-1, a distinct cyclodepsipeptide chemotype, targets a seventh tubulin pharmacological site
Abstract
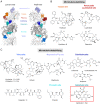
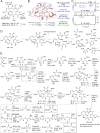
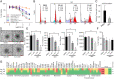
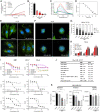
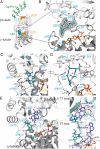
Tubulin-targeted chemotherapy has proven to be a successful and wide spectrum strategy against solid and liquid malignancies. Therefore, new ways to modulate this essential protein could lead to new antitumoral pharmacological approaches. Currently known tubulin agents bind to six distinct sites at α/β-tubulin either promoting microtubule stabilization or depolymerization. We have discovered a seventh binding site at the tubulin intradimer interface where a novel microtubule-destabilizing cyclodepsipeptide, termed gatorbulin-1 (GB1), binds. GB1 has a unique chemotype produced by a marine cyanobacterium. We have elucidated this dual, chemical and mechanistic, novelty through multidimensional characterization, starting with bioactivity-guided natural product isolation and multinuclei NMR-based structure determination, revealing the modified pentapeptide with a functionally critical hydroxamate group; and validation by total synthesis. We have investigated the pharmacology using isogenic cancer cell screening, cellular profiling, and complementary phenotypic assays, and unveiled the underlying molecular mechanism by in vitro biochemical studies and high-resolution structural determination of the α/β-tubulin-GB1 complex.
Keywords: cyanobacteria; marine natural product; microtubules; total synthesis; tubulin.
Conflict of interest statement
Competing interest statement: The University of Florida has filed a patent application relating to the content of this article.
Figures
References
-
- Luesch H., Moore R. E., Paul V. J., Mooberry S. L., Corbett T. H., Isolation of dolastatin 10 from the marine cyanobacterium Symploca species VP642 and total stereochemistry and biological evaluation of its analogue symplostatin 1. J. Nat. Prod. 64, 907–910 (2001). - PubMed
-
- Bai R. L., Pettit G. R., Hamel E., Binding of dolastatin 10 to tubulin at a distinct site for peptide antimitotic agents near the exchangeable nucleotide and vinca alkaloid sites. J. Biol. Chem. 265, 17141–17149 (1990). - PubMed

