Bromodomain proteins regulate human cytomegalovirus latency and reactivation allowing epigenetic therapeutic intervention
- PMID: 33619107
- PMCID: PMC7936348
- DOI: 10.1073/pnas.2023025118
Bromodomain proteins regulate human cytomegalovirus latency and reactivation allowing epigenetic therapeutic intervention
Abstract
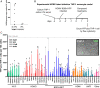
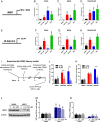
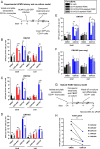
Reactivation of human cytomegalovirus (HCMV) from latency is a major health consideration for recipients of stem-cell and solid organ transplantations. With over 200,000 transplants taking place globally per annum, virus reactivation can occur in more than 50% of cases leading to loss of grafts as well as serious morbidity and even mortality. Here, we present the most extensive screening to date of epigenetic inhibitors on HCMV latently infected cells and find that histone deacetylase inhibitors (HDACis) and bromodomain inhibitors are broadly effective at inducing virus immediate early gene expression. However, while HDACis, such as myeloid-selective CHR-4487, lead to production of infectious virions, inhibitors of bromodomain (BRD) and extraterminal proteins (I-BETs), including GSK726, restrict full reactivation. Mechanistically, we show that BET proteins (BRDs) are pivotally connected to regulation of HCMV latency and reactivation. Through BRD4 interaction, the transcriptional activator complex P-TEFb (CDK9/CycT1) is sequestered by repressive complexes during HCMV latency. Consequently, I-BETs allow release of P-TEFb and subsequent recruitment to promoters via the superelongation complex (SEC), inducing transcription of HCMV lytic genes encoding immunogenic antigens from otherwise latently infected cells. Surprisingly, this occurs without inducing many viral immunoevasins and, importantly, while also restricting viral DNA replication and full HCMV reactivation. Therefore, this pattern of HCMV transcriptional dysregulation allows effective cytotoxic immune targeting and killing of latently infected cells, thus reducing the latent virus genome load. This approach could be safely used to pre-emptively purge the virus latent reservoir prior to transplantation, thereby reducing HCMV reactivation-related morbidity and mortality.
Keywords: I-BET; bromodomain proteins; cytomegalovirus; epigenetics; latency.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Latency-Associated Expression of Human Cytomegalovirus US28 Attenuates Cell Signaling Pathways To Maintain Latent Infection.mBio. 2017 Dec 5;8(6):e01754-17. doi: 10.1128/mBio.01754-17. mBio. 2017. PMID: 29208743 Free PMC article.
-
Transient activation of human cytomegalovirus lytic gene expression during latency allows cytotoxic T cell killing of latently infected cells.Sci Rep. 2016 Apr 19;6:24674. doi: 10.1038/srep24674. Sci Rep. 2016. PMID: 27091512 Free PMC article.
-
Targeting the latent human cytomegalovirus reservoir for T-cell-mediated killing with virus-specific nanobodies.Nat Commun. 2021 Jul 21;12(1):4436. doi: 10.1038/s41467-021-24608-5. Nat Commun. 2021. PMID: 34290252 Free PMC article.
-
Latency and reactivation of human cytomegalovirus.J Gen Virol. 2006 Jul;87(Pt 7):1763-1779. doi: 10.1099/vir.0.81891-0. J Gen Virol. 2006. PMID: 16760381 Review.
-
Sleepless latency of human cytomegalovirus.Med Microbiol Immunol. 2015 Jun;204(3):421-9. doi: 10.1007/s00430-015-0401-6. Epub 2015 Mar 14. Med Microbiol Immunol. 2015. PMID: 25772624 Free PMC article. Review.
Cited by
-
Relationship between the risk of intestinal mucosal Epstein-Barr virus and/or cytomegalovirus infection and peripheral blood NK cells numbers in patients with ulcerative colitis: a cross-sectional study in Chinese population.Front Microbiol. 2024 Dec 4;15:1498483. doi: 10.3389/fmicb.2024.1498483. eCollection 2024. Front Microbiol. 2024. PMID: 39697654 Free PMC article.
-
A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.Nucleic Acids Res. 2024 Feb 28;52(4):1661-1676. doi: 10.1093/nar/gkad1175. Nucleic Acids Res. 2024. PMID: 38084912 Free PMC article.
-
Temporal dynamics of HCMV gene expression in lytic and latent infections.Cell Rep. 2022 Apr 12;39(2):110653. doi: 10.1016/j.celrep.2022.110653. Cell Rep. 2022. PMID: 35417700 Free PMC article.
-
Modeling and Remodeling the Cell: How Digital Twins and HCMV Can Elucidate the Complex Interactions of Viral Latency, Epigenetic Regulation, and Immune Responses.Curr Clin Microbiol Rep. 2023 Sep;10(3):141-151. doi: 10.1007/s40588-023-00201-w. Epub 2023 Jul 29. Curr Clin Microbiol Rep. 2023. PMID: 37901689 Free PMC article.
-
Relevance of BET Family Proteins in SARS-CoV-2 Infection.Biomolecules. 2021 Jul 30;11(8):1126. doi: 10.3390/biom11081126. Biomolecules. 2021. PMID: 34439792 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

