Antibody affinity maturation and plasma IgA associate with clinical outcome in hospitalized COVID-19 patients
- PMID: 33619281
- PMCID: PMC7900119
- DOI: 10.1038/s41467-021-21463-2
Antibody affinity maturation and plasma IgA associate with clinical outcome in hospitalized COVID-19 patients
Abstract
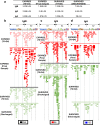
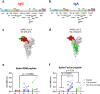
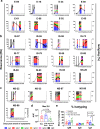
Hospitalized COVID-19 patients often present with a large spectrum of clinical symptoms. There is a critical need to better understand the immune responses to SARS-CoV-2 that lead to either resolution or exacerbation of the clinical disease. Here, we examine longitudinal plasma samples from hospitalized COVID-19 patients with differential clinical outcome. We perform immune-repertoire analysis including cytokine, hACE2-receptor inhibition, neutralization titers, antibody epitope repertoire, antibody kinetics, antibody isotype and antibody affinity maturation against the SARS-CoV-2 prefusion spike protein. Fatal cases demonstrate high plasma levels of IL-6, IL-8, TNFα, and MCP-1, and sustained high percentage of IgA-binding antibodies to prefusion spike compared with non-ICU survivors. Disease resolution in non-ICU and ICU patients associates with antibody binding to the receptor binding motif and fusion peptide, and antibody affinity maturation to SARS-CoV-2 prefusion spike protein. Here, we provide insight into the immune parameters associated with clinical disease severity and disease-resolution outcome in hospitalized patients that could inform development of vaccine/therapeutics against COVID-19.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Johns Hopkins University & Medicine Center for Systems Science and Engineering (CSSE). Coronavirus COVID-19 Global Cases. 2020 [cited 12 May 2020] Available from: https://coronavirus.jhu.edu/map.html. (2020).
-
- Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in china: summary of a report of 72314 cases from the Chinese Center for disease control and prevention. JAMA323, 1239–1242 (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

