In silico detection of SARS-CoV-2 specific B-cell epitopes and validation in ELISA for serological diagnosis of COVID-19
- PMID: 33619344
- PMCID: PMC7900118
- DOI: 10.1038/s41598-021-83730-y
In silico detection of SARS-CoV-2 specific B-cell epitopes and validation in ELISA for serological diagnosis of COVID-19
Abstract
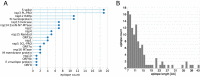
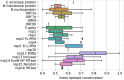
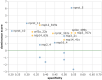
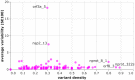
Rapid generation of diagnostics is paramount to understand epidemiology and to control the spread of emerging infectious diseases such as COVID-19. Computational methods to predict serodiagnostic epitopes that are specific for the pathogen could help accelerate the development of new diagnostics. A systematic survey of 27 SARS-CoV-2 proteins was conducted to assess whether existing B-cell epitope prediction methods, combined with comprehensive mining of sequence databases and structural data, could predict whether a particular protein would be suitable for serodiagnosis. Nine of the predictions were validated with recombinant SARS-CoV-2 proteins in the ELISA format using plasma and sera from patients with SARS-CoV-2 infection, and a further 11 predictions were compared to the recent literature. Results appeared to be in agreement with 12 of the predictions, in disagreement with 3, while a further 5 were deemed inconclusive. We showed that two of our top five candidates, the N-terminal fragment of the nucleoprotein and the receptor-binding domain of the spike protein, have the highest sensitivity and specificity and signal-to-noise ratio for detecting COVID-19 sera/plasma by ELISA. Mixing the two antigens together for coating ELISA plates led to a sensitivity of 94% (N = 80 samples from persons with RT-PCR confirmed SARS-CoV-2 infection), and a specificity of 97.2% (N = 106 control samples).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Dahlke C, et al. Distinct early IgA profile may determine severity of COVID-19 symptoms: an immunological case series. medRxiv. 2020 doi: 10.1101/2020.04.14.20059733. - DOI
-
- Hachim A, et al. Beyond the Spike: identification of viral targets of the antibody response to SARS-CoV-2 in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.04.30.20085670. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

