Diet-dependent regulation of TGFβ impairs reparative innate immune responses after demyelination
- PMID: 33619376
- PMCID: PMC7610359
- DOI: 10.1038/s42255-021-00341-7
Diet-dependent regulation of TGFβ impairs reparative innate immune responses after demyelination
Erratum in
-
Author Correction: Diet-dependent regulation of TGFβ impairs reparative innate immune responses after demyelination.Nat Metab. 2023 Mar;5(3):531. doi: 10.1038/s42255-023-00750-w. Nat Metab. 2023. PMID: 36823473 No abstract available.
Abstract
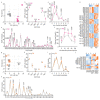
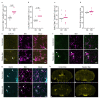
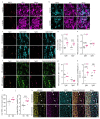
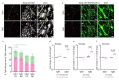
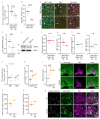
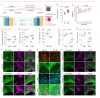
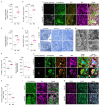
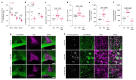
Proregenerative responses are required for the restoration of nervous-system functionality in demyelinating diseases such as multiple sclerosis (MS). Yet, the limiting factors responsible for poor CNS repair are only partially understood. Here, we test the impact of a Western diet (WD) on phagocyte function in a mouse model of demyelinating injury that requires microglial innate immune function for a regenerative response to occur. We find that WD feeding triggers an ageing-related, dysfunctional metabolic response that is associated with impaired myelin-debris clearance in microglia, thereby impairing lesion recovery after demyelination. Mechanistically, we detect enhanced transforming growth factor beta (TGFβ) signalling, which suppresses the activation of the liver X receptor (LXR)-regulated genes involved in cholesterol efflux, thereby inhibiting phagocytic clearance of myelin and cholesterol. Blocking TGFβ or promoting triggering receptor expressed on myeloid cells 2 (TREM2) activity restores microglia responsiveness and myelin-debris clearance after demyelinating injury. Thus, we have identified a druggable microglial immune checkpoint mechanism regulating the microglial response to injury that promotes remyelination.
Conflict of interest statement
G.D.P., K.M.M and J.W.L are paid employees and shareholders of Denali Therapeutics Inc. C.K. is an employee of Lipotype.
Figures







References
-
- Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542:177–185. - PubMed
-
- Hansson GK, Hermansson A. The immune system in atherosclerosis. Nat Immunol. 2011;12:204–212. - PubMed
-
- Franklin RJM, Ffrench-Constant C. Regenerating CNS myelin—from mechanisms to experimental medicines. Nat Rev Neurosci. 2017;18:753–769. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

