A unique subset of glycolytic tumour-propagating cells drives squamous cell carcinoma
- PMID: 33619381
- PMCID: PMC7954080
- DOI: 10.1038/s42255-021-00350-6
A unique subset of glycolytic tumour-propagating cells drives squamous cell carcinoma
Abstract
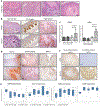
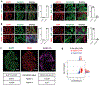
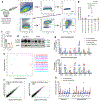
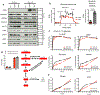
Head and neck squamous cell carcinoma (SCC) remains among the most aggressive human cancers. Tumour progression and aggressiveness in SCC are largely driven by tumour-propagating cells (TPCs). Aerobic glycolysis, also known as the Warburg effect, is a characteristic of many cancers; however, whether this adaptation is functionally important in SCC, and at which stage, remains poorly understood. Here, we show that the NAD+-dependent histone deacetylase sirtuin 6 is a robust tumour suppressor in SCC, acting as a modulator of glycolysis in these tumours. Remarkably, rather than a late adaptation, we find enhanced glycolysis specifically in TPCs. More importantly, using single-cell RNA sequencing of TPCs, we identify a subset of TPCs with higher glycolysis and enhanced pentose phosphate pathway and glutathione metabolism, characteristics that are strongly associated with a better antioxidant response. Together, our studies uncover enhanced glycolysis as a main driver in SCC, and, more importantly, identify a subset of TPCs as the cell of origin for the Warburg effect, defining metabolism as a key feature of intra-tumour heterogeneity.
Conflict of interest statement
Competing Interests Statement
R.M has a financial interest in Galilei Biosciences Inc, a company developing activators of the mammalian SIRT6 protein. R.M.’s interests were reviewed and are managed by MGH and MGB HealthCare in accordance with their conflict of interest policies. N.H. has equity in BioNtech and Related Sciences. N.Y.R.A. is a key opinion leader to Bruker Daltonics. The other authors declare no competing interests.
Figures








References
-
- Visvader JE & Lindeman GJ Cancer Stem Cells: Current Status and Evolving Complexities. Cell Stem Cell 10, 717–728 (2012). - PubMed
-
- Malanchi I et al. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature 452, 650–653 (2008). - PubMed
-
- Boumahdi S et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 10, 246–250 (2014). - PubMed
REFERENCES for METHODS
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

