Multiple cell types contribute to the atherosclerotic lesion fibrous cap by PDGFRβ and bioenergetic mechanisms
- PMID: 33619382
- PMCID: PMC7905710
- DOI: 10.1038/s42255-020-00338-8
Multiple cell types contribute to the atherosclerotic lesion fibrous cap by PDGFRβ and bioenergetic mechanisms
Abstract
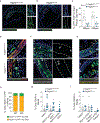
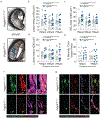
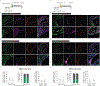
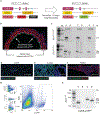
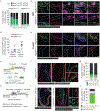
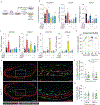
Stable atherosclerotic plaques are characterized by a thick, extracellular matrix-rich fibrous cap populated by protective ACTA2+ myofibroblast (MF)-like cells, assumed to be almost exclusively derived from smooth muscle cells (SMCs). Herein, we show that in murine and human lesions, 20% to 40% of ACTA2+ fibrous cap cells, respectively, are derived from non-SMC sources, including endothelial cells (ECs) or macrophages that have undergone an endothelial-to-mesenchymal transition (EndoMT) or a macrophage-to-mesenchymal transition (MMT). In addition, we show that SMC-specific knockout of the Pdgfrb gene, which encodes platelet-derived growth factor receptor beta (PDGFRβ), in Apoe-/- mice fed a Western diet for 18 weeks resulted in brachiocephalic artery lesions nearly devoid of SMCs but with no changes in lesion size, remodelling or indices of stability, including the percentage of ACTA2+ fibrous cap cells. However, prolonged Western diet feeding of SMC Pdgfrb-knockout mice resulted in reduced indices of stability, indicating that EndoMT- and MMT-derived MFs cannot compensate indefinitely for loss of SMC-derived MFs. Using single-cell and bulk RNA-sequencing analyses of the brachiocephalic artery region and in vitro models, we provide evidence that SMC-to-MF transitions are induced by PDGF and transforming growth factor-β and dependent on aerobic glycolysis, while EndoMT is induced by interleukin-1β and transforming growth factor-β. Together, we provide evidence that the ACTA2+ fibrous cap originates from a tapestry of cell types, which transition to an MF-like state through distinct signalling pathways that are either dependent on or associated with extensive metabolic reprogramming.
Conflict of interest statement
Competing Interest Statement:
AF declares one competing interest with Amgen Inc. All other authors declare no competing interests.
Figures








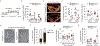


References
-
- Virmani Renu, Kolodgie Frank D., Burke Allen P., Farb Andrew & Schwartz Stephen M. Lessons From Sudden Coronary Death. Arteriosclerosis, Thrombosis, and Vascular Biology 20, 1262–1275 (2000). - PubMed
-
- Libby P, Ridker PM & Hansson GK Progress and challenges in translating the biology of atherosclerosis. Nature 473, 317–325 (2011). - PubMed
-
- Feil S et al. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ. Res 115, 662–667 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

