Tetracycline as an inhibitor to the SARS-CoV-2
- PMID: 33619758
- PMCID: PMC8014839
- DOI: 10.1002/jcb.29909
Tetracycline as an inhibitor to the SARS-CoV-2
Abstract
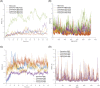
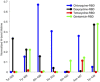
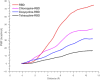
The coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remains an extant threat against public health on a global scale. Cell infection begins when the spike protein of SARS-CoV-2 binds with the human cell receptor, angiotensin-converting enzyme 2 (ACE2). Here, we address the role of tetracycline as an inhibitor for the receptor-binding domain (RBD) of the spike protein. Targeted molecular investigation show that tetracycline binds more favorably to the RBD (-9.40 kcal/mol) compared to doxycycline (-8.08 kcal/mol), chloroquine (-6.31 kcal/mol), or gentamicin (-4.83 kcal/mol) while inhibiting attachment to ACE2 to a greater degree (binding efficiency of 2.98 kcal/(mol nm2 ) for tetracycline-RBD, 5.16 kcal/(mol nm2 ) for doxycycline-RBD, 5.59 kcal/(mol nm2 ) for chloroquine-RBD, and 7.02 kcal/(mol nm2 ) for gentamicin-RBD. Stronger inhibition by tetracycline is verified with nonequilibrium PMF calculations, for which the tetracycline-RBD complex exhibits the lowest free energy profile along the dissociation pathway from ACE2. Tetracycline binds to tyrosine and glycine residues on the viral contact interface that are known to modulate molecular recognition and bonding affinity. These RBD residues also engage in significant hydrogen bonding with the human receptor ACE2. The ability to preclude cell infection complements the anti-inflammatory and cytokine suppressing capability of tetracycline; this may reduce the duration of ICU stays and mechanical ventilation induced by the coronavirus SARS-CoV-2.
Keywords: COVID-19; Coronavirus; SARS-CoV-2; tetracycline.
© 2021 Wiley Periodicals LLC.
Figures

References
-
- Muhammad A, Ghazanfar A. Docking study of chloroquine and hydroxychloroquine interaction with rna binding domain of nucleocapsid phospho‐protein—an in silico insight into the comparative efficacy of repurposing antiviral drugs. J Biomol Struct Dyn. 2020:1‐13. 10.1080/07391102.2020.1775703 - DOI - PubMed
-
- Sekiou O, Bouziane I, Bouslama Z, Djemel A. In‐silico identification of potent inhibitors of COVID‐19 main protease (Mpro) and angiotensin converting enzyme 2 (ACE2) from natural products: quercetin, hispidulin, and cirsimaritin exhibited better potential inhibition than hydroxy‐chloroquine against COVID‐19 main protease active site and ACE2. 2020. 10.26434/chemrxiv.12181404 - DOI
-
- Byrne JD, Shakur R, Collins J, et al. Prophylaxis with tetracyclines in ards: potential therapy for covid‐19‐induced ards? medRxiv. 2020. 10.1101/2020.07.22.20154542 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

