Mapping the plant proteome: tools for surveying coordinating pathways
- PMID: 33620075
- PMCID: PMC8166341
- DOI: 10.1042/ETLS20200270
Mapping the plant proteome: tools for surveying coordinating pathways
Abstract
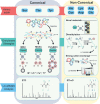
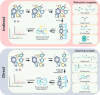
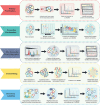
Plants rapidly respond to environmental fluctuations through coordinated, multi-scalar regulation, enabling complex reactions despite their inherently sessile nature. In particular, protein post-translational signaling and protein-protein interactions combine to manipulate cellular responses and regulate plant homeostasis with precise temporal and spatial control. Understanding these proteomic networks are essential to addressing ongoing global crises, including those of food security, rising global temperatures, and the need for renewable materials and fuels. Technological advances in mass spectrometry-based proteomics are enabling investigations of unprecedented depth, and are increasingly being optimized for and applied to plant systems. This review highlights recent advances in plant proteomics, with an emphasis on spatially and temporally resolved analysis of post-translational modifications and protein interactions. It also details the necessity for generation of a comprehensive plant cell atlas while highlighting recent accomplishments within the field.
Keywords: plant proteins; post translational modification; protein–protein interactions; proteomics.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
Similar articles
-
Plant Proteomics and Systems Biology.Adv Exp Med Biol. 2021;1346:51-66. doi: 10.1007/978-3-030-80352-0_3. Adv Exp Med Biol. 2021. PMID: 35113395
-
Navigating the landscape of plant proteomics.J Integr Plant Biol. 2025 Mar;67(3):740-761. doi: 10.1111/jipb.13841. Epub 2025 Jan 15. J Integr Plant Biol. 2025. PMID: 39812500 Review.
-
Advances in functional proteomics to study plant-pathogen interactions.Curr Opin Plant Biol. 2021 Oct;63:102061. doi: 10.1016/j.pbi.2021.102061. Epub 2021 Jun 5. Curr Opin Plant Biol. 2021. PMID: 34102449 Review.
-
Exploring the diversity of plant proteome.J Integr Plant Biol. 2021 Jul;63(7):1197-1210. doi: 10.1111/jipb.13087. Epub 2021 Apr 1. J Integr Plant Biol. 2021. PMID: 33650765 Review.
-
Plant Proteome Dynamics.Annu Rev Plant Biol. 2022 May 20;73:67-92. doi: 10.1146/annurev-arplant-102620-031308. Epub 2022 Feb 9. Annu Rev Plant Biol. 2022. PMID: 35138880 Review.
Cited by
-
Translational profile of coding and non-coding RNAs revealed by genome wide profiling of ribosome footprints in grapevine.Front Plant Sci. 2023 Feb 8;14:1097846. doi: 10.3389/fpls.2023.1097846. eCollection 2023. Front Plant Sci. 2023. PMID: 36844052 Free PMC article.
-
Detergent-Assisted Protein Digestion-On the Way to Avoid the Key Bottleneck of Shotgun Bottom-Up Proteomics.Int J Mol Sci. 2022 Nov 11;23(22):13903. doi: 10.3390/ijms232213903. Int J Mol Sci. 2022. PMID: 36430380 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
